Abstract
The innate immune system relies on evolutionally conserved Toll-like receptors (TLRs) to recognize diverse microbial molecular structures. Most TLRs depend on a family of adaptor proteins termed MyD88s to transduce their signals. Critical roles of MyD88-1–4 in host defense were demonstrated by defective immune responses in knockout mice. In contrast, the sites of expression and functions of vertebrate MyD88-5 have remained elusive. We show that MyD88-5 is distinct from other MyD88s in that MyD88-5 is preferentially expressed in neurons, colocalizes in part with mitochondria and JNK3, and regulates neuronal death. We prepared MyD88-5/GFP transgenic mice via a bacterial artificial chromosome to preserve its endogenous expression pattern. MyD88-5/GFP was detected chiefly in the brain, where it associated with punctate structures within neurons and copurified in part with mitochondria. In vitro, MyD88-5 coimmunoprecipitated with JNK3 and recruited JNK3 from cytosol to mitochondria. Hippocampal neurons from MyD88-5–deficient mice were protected from death after deprivation of oxygen and glucose. In contrast, MyD88-5–null macrophages behaved like wild-type cells in their response to microbial products. Thus, MyD88-5 appears unique among MyD88s in functioning to mediate stress-induced neuronal toxicity.
MyD88 (myeloid differentiation primary response gene 88) is the founding member of a family of mammalian cytosolic adaptor proteins distinguished by a C-terminal Toll-interleukin 1 receptor (TIR) domain. The four best-studied members of the MyD88 family all play prominent roles in innate immunity. MyD88 is a critical intermediary in signaling through interleukin-1 receptors, interleukin-18 receptor, and most Toll-like receptors (TLRs), except for TLR3 (1, 2). MyD88-2, which is also known as TIR domain–containing adaptor protein (TIRAP) or MyD88 adaptor–like (MAL), is required for TLR4- and MyD88-dependent responses and for signaling via TLR2 (3–5). MyD88-3, which is commonly called TIR domain–containing adaptor inducing interferon-β (TRIF) or TIR domain–containing adaptor molecule-1 (TICAM-1), mediates responses to ligation of TLR3, as well as those responses to ligation of TLR4 that are MyD88-independent (6–8). MyD88-4, which is also called TRIF-related adaptor molecule (TRAM) or TIR-domain containing protein (TIRP), shares with MyD88-3 the mediation of TLR4-dependent, but MyD88-independent responses (9–11).
In contrast, the function of MyD88-5, which is also called sterile α and HEAT/Armadillo motifs containing protein (SARM), remains a mystery (12). That the function of MyD88-5 is fundamental is suggested by its high degree of conservation among fly, worm, and mammals. That MyD88-5 plays a different role than the other members of the MyD88 family is suggested by its 7–8 N-terminal HEAT/Armadillo repeats and two sterile α motif (SAM) domains, structures typically involved in protein–protein interactions in cytoskeletal regulation and intracellular signaling (13–15).
In Caenorhabditis elegans, the MyD88-5 homologue TIR-1 is the sole cytoplasmic protein with a TIR domain. TIR-1 was hypothesized to mediate signaling from TOL-1, which is the only TLR homologue. Indeed, knockdown of tir-1 shortened the lifespan of worms feeding on Drechmeria coniospora, Serratia marcescens (16), Pseudomonas aeruginosa, and Enterococcus faecalis (17). However, this effect was independent of TOL-1 (16). Thus, rather than mediating signaling from TOL-1, TIR-1 may have controlled recognition of environmental cues derived from the pathogenic bacteria. Consistent with the latter interpretation, tir-1 mutants displayed bilateral, symmetric expression of an olfactory receptor candidate gene, str-2, in both of their olfactory neurons (18). In contrast, in wild-type worms, only one of the two neurons expresses this gene (19–21); the asymmetry may contribute to the worm's ability to mount an adaptive directional response to environmental cues. Epistasis analysis suggested that TIR-1 may influence olfactory neuronal development at a step between UNC-43, a calmodulin-dependent protein kinase II homologue, and NSY-1, a homologue of human ASK1, which is a MAP kinase kinase kinase (18, 22).
In mammals, the roles of MyD88-1–4 were demonstrated by their ability to activate NF-κB or the IFN-β promoter when overexpressed and by a loss of TLR-mediated function when individual MyD88 members were target-deleted (6, 10, 11, 23, 24). In contrast, overexpression of MyD88-5 in HEK293 cells did not induce expression of NF-κB– or IFN-β–regulated reporter genes (17, 25), and MyD88-5 knockout mice have not previously been described. Recently, Carty et al. (26) suggested that MyD88-5 might serve as a negative regulator of TRIF-dependent TLR signaling based primarily on overexpression or siRNA-mediated suppression of MyD88-5 in HEK 293 cells. It is not clear, however, whether myeloid cells express MyD88-5.
To learn the role of mammalian MyD88-5, we cloned the mouse and human genes, raised antibodies to the recombinant proteins, and performed expression, transfection, colocalization, coimmunoprecipitation, bacterial artificial chromosome (BAC) transgenic, and knockout mouse studies. These approaches revealed that human and mouse myeloid cells expressed little MyD88-5. Mouse macrophages responded to poly(I:C) and LPS normally in the absence of the MyD88-5 gene. In contrast, MyD88-5 is expressed primarily in neurons, where it associates with mitochondria, microtubules, and JNK3 and regulates neuronal death during deprivation of oxygen and glucose.
RESULTS
MyD88-5 is evolutionarily conserved
Proteins with a TIR domain are found throughout the plant and animal kingdoms, with or without transmembrane domains; MyD88 is an example of the latter subset (27–29). Shortly after it was found that not all TLR-dependent innate immune responses depend on MyD88 (30), we conducted a bioinformatic search for additional proteins with TIR domains that like MyD88, but in contrast to TLRs, were predicted to be cytoplasmic. Sequence homology led only to identification of MyD88-2 (3–5, 24), -3 (6, 8), and -4 (9, 11), each of which was independently described during our analysis. However, when we searched SMART and PFAM databases, which are based on position-specific scoring matrix systems, we identified MyD88-5 as a protein with two SAM domains flanked by a C-terminal TIR domain and N-terminal HEAT/Armadillo repeats (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20070868/DC1). While this functional characterization was in progress, cloning of MyD88-5 was reported (12). MyD88-5 homologues are found in flies, worms, fish, and mammals. In its phylogenetic context (Fig. S1 B), there is a deep division of MyD88-5 homologues of mammals, Drosophila melanogaster (Ect-4) (31), and C. elegans (TIR-1) from other TIRPs, suggesting that the functions of MyD88-5s may differ from those of the IL-1/18 receptor family, the TLRs, the Tolls, and the other MyD88s.
MyD88-5 is expressed mainly in brain
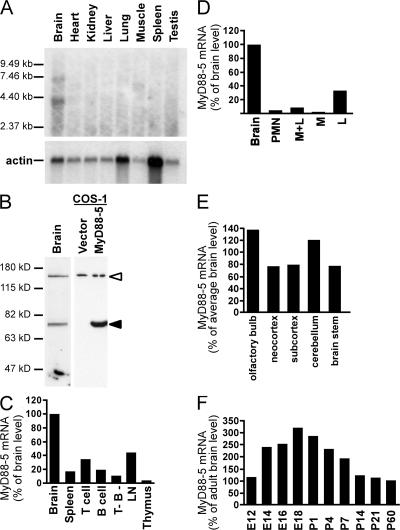
Northern blot analysis using part of the cloned mouse MyD88-5 cDNA containing both SAM domains and the TIR domain as a probe revealed the expected ∼7.4-kb transcript in brain, but not in heart, kidney, liver, lung, skeletal muscle, spleen, or thymus (Fig. 1 A), in contrast to the study by Mink et al. (12). An additional transcript of shorter size (∼4 kb) may represent a splice variant, but antibody raised against full-length recombinant mouse MyD88-5 detected only one polypeptide in mouse brain and in MyD88-5–transfected COS-1 cells, and its apparent molecular weight (79 kD) matched that expected for the full-length protein (Fig. 1 B).
Figure 1.
MyD88-5 is expressed mainly in brain, but not in myeloid cells. (A) Northern blot analysis of MyD88-5 expression in mouse tissues using the two SAM domains and part of the TIR domain (1,200–1,816 bp) as a probe. An actin probe was used as a loading control. (B) Western blot with chicken antibody raised against full-length recombinant mouse MyD88-5 reveals a 79-kD polypeptide in mouse brain (left lane) and in COS-1 cells transfected with a Flag-tagged MyD88-5 construct (right lane, filled arrowhead), but not in vector-transfected COS-1 cells (center lane). The open arrowhead indicates a nonspecific band. (C) Relative quantitation of MyD88-5 transcripts in mouse tissues and cells. Expression was highest in brain and moderate in lymph node (LN) and purified splenic T cells, but was barely detectable in splenic populations (chiefly macrophages) depleted of T and B cells (T- B-). Data are quantitative real-time PCR results normalized to mouse GAPDH and expressed as the percentage of the level in brain. One of three independent experiments is shown. (D) Relative quantitation of MyD88-5 transcripts in human tissues and cells. Expression of human MyD88-5 mRNA is highest in brain and detectable in lymphocytes (L). Polymorphonuclear leukocytes (PMN), monocytes (M), and lymphocytes were purified from human blood. Data are expressed as in C for one of three independent experiments. (E) MyD88-5 expression in various anatomic regions of adult mouse brain. Data are expressed as in C, and are from one of two independent experiments. (F) MyD88-5 expression in mouse brain during development. E12-18, embryonic days after conception; P1-60, postnatal days. qRT-PCR results are normalized to GAPDH and shown as the percentage of MyD88-5 expression in adult brain.
Quantitative real-time RT-PCR (qRT-PCR) confirmed the preferential expression of MyD88-5 mRNA in brain of mouse (Fig. 1 C) and human (Fig. 1 D). MyD88-5 transcripts were also detected in mouse spleen and lymph nodes, but not in thymus (Fig. 1 C). In human peripheral blood, lymphocyte-enriched populations expressed MyD88-5 mRNA, whereas monocyte- and PMN-enriched populations did not (Fig. 1 D). Focusing on brain as the richest source of MyD88-5, we compared transcript levels in different anatomic regions. Expression levels were comparable across the organ (Fig. 1 E). During embryonic development of the mouse, MyD88-5 expression increased gradually in the brain, peaking on day 18, which is a period of massive neuronal proliferation and programmed cell death (32). Levels fell threefold after birth (Fig. 1 F).
MyD88-5 is expressed in neurons
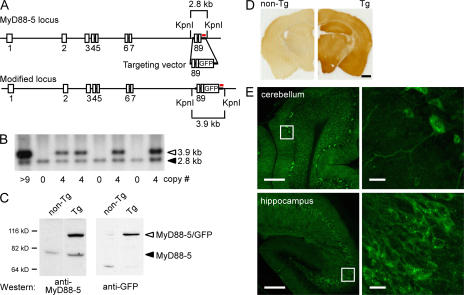
Next, we sought to identify the cell types responsible for the relatively high level of MyD88-5 expression in brain. Despite extensive efforts to raise antibodies in rabbits and chickens against recombinant mouse and human MyD88-5, the resulting reagents sufficed only for Western blots, not for immunoprecipitation, immunohistochemistry, or flow cytometry. As an alternative strategy, we generated enhanced GFP-tagged MyD88-5 transgenic lines of mice using a BAC. RP23-399H5, which is a 196-kb BAC clone, included an extra 156 kb of mouse genomic DNA upstream of the MyD88-5 translation start site, as well as 14 kb after the stop codon. This configuration was expected to include the full complement of endogenous promoter/enhancer sequences and to allow for physiologic generation of alternative splice variants. Therefore, the expression of the transgene was expected to faithfully reflect the expression of the endogenous gene (33). As schematized in Fig. 2 A, we engineered Myd88-5 to be expressed with a C-terminal GFP fusion construct. Southern blot of a KpnI restriction enzyme digest (Fig. 2 B) showed the integrated DNA in a band migrating at 3.9 kb and the endogenous gene at 2.8 kb. Founders with one integrated copy of the BAC plasmid did not show a detectable level of GFP expression by fluorescence or anti-GFP Western blot. Therefore, we established transgenic lines from founders expressing 4–12 copies of the transgene, but for unknown reasons, only mice with 4 copies of the transgene produced F1 progeny. Western blot analysis with anti–MyD88-5 antibody detected both the MyD88-5/GFP fusion protein and the endogenous MyD88-5 in the brains of three independent founders, as well as in three independent F1 generations derived from another set of founders, and Western blot analysis with anti-GFP antibody confirmed the expression of the fusion protein in the brain (Fig. 2 C). FACS analysis detected no expression of MyD88-5/GFP in thymocytes or CD11b+ splenocytes (macrophages) and low-level expression in CD3+ splenocytes (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070868/DC1), which is consistent with the expression pattern of endogenous MyD88-5 transcripts (Fig. 1 C).
Figure 2.
Generation of transgenic mice expressing MyD88-5-GFP fusion protein and its expression in neurons in the brain. (A) Schematic of the MyD88-5 locus in BAC clone RP23-399H5, the targeting vector, and the predicted structure of the modified MyD88-5 locus. Exon numbers are indicated. Probe used for Southern blotting is shown in red. Restriction sites and predicted fragment sizes are indicated. The stop codon in exon 9 is removed, and GFP is introduced in-frame. (B) Representative Southern blot of restricted genomic DNA from transgenic (Tg) and nontransgenic mice. Transgene copy numbers shown below are calculated by comparing the intensity of the endogenous band (filled arrowhead) and transgenic band (open arrowhead). (C) Expression of endogenous MyD88-5 and MyD88-5/GFP fusion protein in transgenic mouse brain. (left) Western blot with anti–MyD88-5 antibody. (right) Western blot with anti-GFP antibody. The fusion protein has the expected molecular weight of 105 kD. (D) Widespread expression of MyD88-5/GFP in the brain of transgenic mice. Brain sections from a nontransgenic mouse (left) and a transgenic (right) mouse were stained together with anti-GFP antibody. (E) Neuronal expression of MyD88-5/GFP. Fluorescent confocal microscopy captured direct GFP signals from unstained brain sections from a transgenic founder mouse. Right images are magnifications of the insets from the left. Bars: (D) 1 mm; (E, left) 200 μm; (E, right) 20 μm.
Next, using anti-GFP antibody for immunohistochemistry, we compared brain sections from transgenic mice and their nontransgenic littermates. Expression of GFP was widespread throughout the brain of the transgenics (Fig. 2 D), which is consistent with the widespread cerebral and cerebellar expression of endogenous MyD88-5 transcripts (Fig. 1 E).
In a transgenic mouse that had integrated 12 copies of the transgene, the fusion protein was readily detected via the intrinsic fluorescence of GFP. Confocal microscopic images of GFP fluorescence obtained without antibody amplification showed expression solely in neurons and not in oligodendrocytes, astrocytes, microglia, meningeal lining cells, or cells of the vasculature. Expression was particularly robust in the pyramidal cells of the hippocampus (CA1-3 cells) and Purkinje cells of the cerebellum. The MyD88-5 fusion protein appeared to be exclusively cytoplasmic in localization and partly punctate in distribution (Fig. 2 E).
MyD88-5 is associated with mitochondria
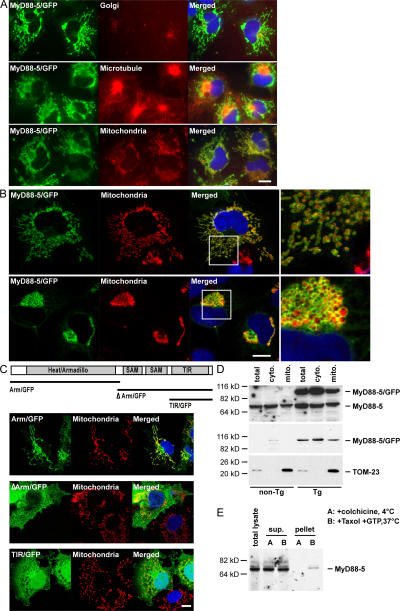
To identify the punctate structures with which much of the MyD88-5 fusion protein appeared to associate, we transfected COS-1 cells with a MyD88-5/GFP fusion construct. The cells were costained with anti–γ-adaptin for visualization of Golgi, anti-tubulin for microtubules, or MitoTracker Red CMXRos for mitochondria. MyD88-5/GFP fusion protein strongly colocalized with mitochondria and partly colocalized with microtubules (Fig. 3 A), structures along which mitochondria move (34). Confocal images suggested that MyD88-5 did not reside within the mitochondrial matrix, where the MitoTracker Red CMXRos dye accumulates, but instead was localized outside the mitochondrial matrix (Fig. 3 B, right), which is consistent with the absence of a mitochondrial targeting sequence in MyD88-5.
Figure 3.
Mitochondrial association of MyD88-5. (A) Selective colocalization of MyD88-5/GFP fusion protein with mitochondria and microtubules. COS-1 cells were transfected with a MyD88-5/GFP fusion construct. 18 h after the transfection, cells were stained with MitoTracker Red CMXRos for 20 min and fixed, or fixed, permeabilized, and stained with antibody for Golgi (anti–γ-adaptin) or microtubules (anti-tubulin). (B) Extrinsic association of MyD88-5 with mitochondria. Confocal microscopic images of COS-1 cells transfected with MyD88-5/GFP and stained with MitoTracker Red CMXRos. (top) Cells expressing lower levels of MyD88-5; (bottom) cells expressing higher levels of MyD88-5, with effects on mitochondrial shape and localization. (right) Magnified images of the boxed areas from the images on the left. (C) Analysis of domains of MyD88-5 contributing to subcellular localization and mitochondrial association. COS-1 cells were transfected with the indicated constructs. 18 h later, mitochondria were stained with MitoTracker Red CMXRos before fixation. (D) Partial copurification of endogenous MyD88-5 and transgenic MyD88-5/GFP with mitochondria from brain. Mitochondria were isolated from wild type (non-Tg) or transgenic (Tg) mouse brains by differential centrifugation and lysates were Western blotted with antibody to MyD88-5 (top), GFP (middle), or TOM-23 (bottom). TOM-23 is an intrinsic mitochondrial membrane protein whose relative abundance serves as a loading control. (E) Copurification of endogenous MyD88-5 and microtubules from mouse brain. Microtubules were either polymerized with Taxol and GTP at 37°C or prevented from polymerization with colchicine at 4°C before purification by sucrose gradient centrifugation. Purified microtubule fractions (pellet), unpolymerized fractions (sup), and total brain lysate (total lysate) were Western blotted with antibody to MyD88-5. Bars, 10 μm.
The distribution and shape of mitochondria varied in COS-1 cells depending on their expression level of MyD88-5. In cells expressing a low to moderate level of MyD88-5, mitochondria were small, rod-shaped, and distributed widely throughout the cytoplasm, as in untransfected cells (Fig. 3 B, top). In cells with high levels of MyD88-5 expression, mitochondria were large, round, and clustered around the microtubule-organizing center, being nearly absent from the periphery (Fig. 3 B, bottom). No changes were noted in the appearance or distribution of other organelles, such as the Golgi apparatus or microtubules (Fig. 3 A). Thus, overexpression of MyD88-5 may have disrupted the association of mitochondria with microtubules.
We then attempted to assign the mitochondria-binding and mitochondria-redistributing functions of MyD88-5 to specific domains by transfecting COS-1 cells with constructs expressing truncated forms of MyD88-5. MyD88-5 lacking the two SAM domains and the TIR domain (Arm/GFP) localized to mitochondria, but did not make them cluster (Fig. 3 C, top). A small fraction of this mutant protein was also detected in nuclei. MyD88-5 lacking the HEAT/Armadillo repeats (ΔArm/GFP) did not localize to mitochondria, but was instead distributed evenly in the cytoplasm, sparing the nuclei (Fig. 3 C, middle). In contrast, a construct consisting of only the TIR domain (TIR/GFP) distributed evenly throughout the cytoplasm and the nuclei (Fig. 3 C, bottom). Thus, HEAT/Armadillo repeats control the association of MyD88-5 with mitochondria, whereas either or both of the SAM domains and the TIR domain are required for mitochondrial clustering.
The apparent association of MyD88-5 with mitochondria in transfected cells in vitro was confirmed biochemically in nontransfected cells in vivo by isolating mitochondrial fractions from mouse brain. MyD88-5 was found both in the mitochondrial fraction and in the soluble fraction (Fig. 3 D), which is consistent with the morphologic picture of a peripheral rather than integral association of MyD88-5 with mitochondria. A peripheral association of MyD88-5 with mitochondria might permit much of the protein to dissociate from mitochondria during the extensive isolation procedure.
Likewise, the apparent morphologic association of MyD88-5 with microtubules in transfected cells was confirmed biochemically in nontransfected cells in vivo by isolating microtubule fractions from mouse brain. MyD88-5 cosedimented with polymerized microtubules, but not when polymerization of tubulin was blocked by colchicine (Fig. 3 E).
MyD88-5 associates with JNK3 and recruits it to the mitochondrial compartment
The InteractomeDB w15 database (http://vidal.dfci.harvard.edu/interactomedb/i-View/) catalogs the results of a high-throughput yeast two-hybrid screen used for systematic identification of protein–protein interactions in C. elegans (35, 36). According to the database, the MyD88-5 worm homologue TIR-1 binds to at least seven proteins beside itself, most of which have mammalian homologues whose isoforms are expressed in brain. One is a homologue of JNK3 (MAPK10 isoform-3). We focused on the possibility that JNK3 may be a MyD88-5 binding partner because of the reported association of JNKs with mitochondria (37) and microtubules (38, 39). Interest in the candidacy of JNK3 was heightened further by its important role in regulating neuronal death during stress (40–46).
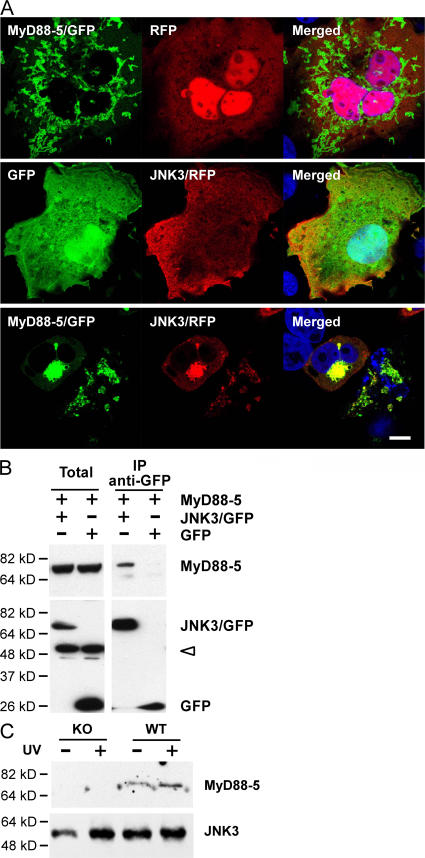
Accordingly, we transfected COS-1 cells with a JNK3/red fluorescent protein (RFP) fusion protein. JNK3 localized mainly to the plasma membrane and cytoplasm (Fig. 4 A, middle). When JNK3 was coexpressed with a MyD88-5/GFP fusion protein, the distribution of JNK3 changed dramatically; the majority of the protein appeared at the mitochondria (Fig. 4 A, bottom). The redistribution was not caused by the RFP fusion tag, because RFP alone did not localize to mitochondria when expressed with MyD88-5/GFP (Fig. 4 A, top).
Figure 4.
MyD88-5–dependent recruitment of JNK3 to mitochondria. (A) Redistribution of JNK3 to mitochondria in the presence of MyD88-5. COS-1 cells were transfected with MyD88-5/GFP and RFP (top), GFP and JNK3/RFP (middle), or MyD88-5/GFP and JNK3/RFP (bottom). Mitochondria were stained with MitoTracker Red CMXRos. Bar, 10 μm. (B) Coimmunoprecipitation of MyD88-5 with JNK3/GFP, but not with GFP alone, from transfected COS-1 cells. Immunoprecipitation was with anti-GFP antibody and protein G–Sepharose and Western blotting was with antibody to MyD88-5 (top) or GFP (bottom). Open arrowhead indicates nonspecific bands. (C) Coimmunoprecipitation of MyD88-5 with JNK3 in hippocampal slices from wild-type (WT) or MyD88-5 knockout (KO) mice. Half of the brain slices were preexposed to 120 Joules/m2 of UV light. Immunoprecipitation was done with anti-JNK1,3 and protein G–Sepharose and Western blot with anti-JNK3 and anti-MyD88-5.
The microscopic association of MyD88-5 with JNK3 was confirmed biochemically. MyD88-5 could be coimmunoprecipitated with JNK3/GFP by anti-GFP antibody in COS-1 cells cotransfected with MyD88-5 and JNK3/GFP fusion constructs (Fig. 4 B). To confirm this association in a more physiologically relevant context, we sliced freshly isolated hippocampi into 150-μm-thick sections and exposed them to 120 Joules/m2 of UV light. We precipitated JNK3 and MyD88-5 together from these hippocampal lysates with antibody against JNK3 (Fig. 4 C). This association was more pronounced after UV irradiation, suggesting that MyD88-5 may be associated more avidly with activated JNK3. As a control, we used hippocampal slides from MyD88-5 knockout mice, whose generation is described in the next section.
Together, these data support the speculation that MyD88-5 acts as a scaffold not only to connect mitochondria to microtubules but also to bring JNK3 to mitochondria.
Hippocampal neurons from MyD88-5–deficient mice are protected from death when deprived of oxygen and glucose
To probe the function of MyD88-5, we took a clue from its association with mitochondria and JNK3, both of which regulate apoptosis during cellular stress. One profound stress on the brain of major clinical significance is cerebral ischemia, which results in the deprivation of oxygen and glucose during vascular insufficiency, such as cerebral stroke. With ischemia-reperfusion injury, JNKs and BAD are increased in hippocampal neurons and translocate to the outer mitochondrial membrane (40, 41). JNK3-null mice are protected from hippocampal cell death induced by cerebral ischemia (43, 44). Thus, we hypothesized that genetic deletion of MyD88-5 by decreasing the association of JNK3 with mitochondria would partially mimic JNK3 deficiency and protect neurons from death during deprivation of oxygen and glucose.
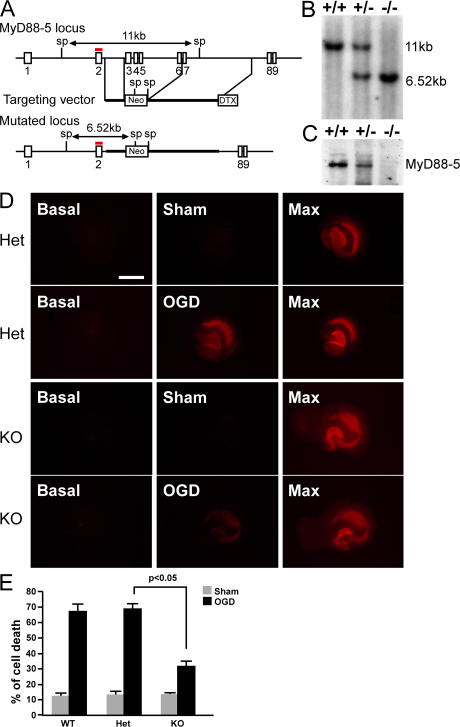
To test this hypothesis, we generated Myd88-5–null mice using the strategy for homologous recombination depicted in Fig. 5 A. Exons 3–6 were replaced with a neomycin resistance gene in reverse orientation such that any read-through from exon 2–7 would be out of frame, precluding production of a truncated form of MyD88-5. Production of wild-type, hemizygous-deficient, and null mice was confirmed by Southern (Fig. 5 B) and Western blot (Fig. 5 C). Null pups were born at the expected Mendelian frequency from hemizygous parents. Whole-body pathological evaluation at gross and microscopic levels revealed no abnormalities. In particular, the brains of MyD88-5–null mice appeared morphologically normal, as was the case for JNK3-null mice (46).
Figure 5.
Generation of MyD88-5 knockout mice and protection of their hippocampal neurons from OGD-induced cell death. (A) Schematic of the MyD88-5 locus, targeting vector, and predicted structure of the mutated locus. Exon numbers are indicated. The probe used for Southern blotting is shown in red. Restriction enzyme sites and predicted sizes of the digests are shown. Sp, SphI; Neo, neomycin resistance gene; DTX, diphtheria toxin gene. (B) Southern blot analysis of genomic DNA from MyD88-5 wild-type, hemizygous-deficient, and null mice. (C) Western blot analysis of MyD88-5 expression in wild-type, hemizygous-deficient, and null mice. 50 μg of brain lysates were subjected to SDS-PAGE. Affinity-purified chicken anti–MyD88-5 antibody was used for detection. (D) Propidium iodide staining of dead neurons in hippocampal slices at the start of the OGD period (basal), after OGD (OGD), or the same period without OGD treatment (sham), and after exposure to 1 mM NMDA to induce a maximal cell death (Max). Hippocampi illustrated were from MyD88-5+/− (Het) or MyD88-5−/− mice (KO). Bar, 1 mm. (E) Quantitative evaluation of cell death after sham or OGD treatment in MyD88-5+/+ (sham, n = 6; OGD, n = 13) MyD88-5+/− (sham, n = 11; OGD, n = 22) and MyD88-5−/− (sham, n = 5; OGD, n = 29) slices. P < 0.05, Student's t test.
A standard model for oxygen and glucose deprivation (OGD) uses hippocampal slices in vitro (47), in which neuronal viability is assessed by propidium iodide (PI) staining before and after OGD. In MyD88-5+/+ and MyD88-5+/− mice, OGD induced massive cell death in the CA1 region of the hippocampi (Fig. 5, D and E). Compared with MyD88-5+/−, the cell death induced by OGD in the MyD88-5−/− hippocampi was reduced from 67.21 ± 5.10 to 31.89 ± 3.34% (P < 0.05).
To test whether ablation of MyD88-5 affects TLR-mediated signaling pathways, we isolated bone marrow–derived macrophages from MyD88-5–deficient mice and their wild-type littermates. In response to LPS, poly IC, Pam3Cys, or CpG macrophages with or without an intact MyD88-5 gene expressed TNF and MCP-1 at comparable levels (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070868/DC1). These results suggest that deletion of MyD88-5 neither enhances nor suppresses macrophage responses to stimuli that trigger TLR-2–4 or -9.
DISCUSSION
MyD88-5 is the only TIR-domain containing cytoplasmic protein conserved from worm to mammals and the only MyD88 family member found in C. elegans. The prominent contribution of all other known members of the MyD88 family to innate immunity fueled the expectation that MyD88-5 would play a similar role. However, C. elegans' MyD88-5 does not appear to mediate signaling from C. elegans' TLR (16). Overexpression of human MyD88-5 failed to activate NF-κB– or IFN-β–regulated reporter genes (17, 25). Carty et al. (26) reported that MyD88-5 interacts with TRIF and negatively regulates TRIF-mediated LPS and poly(I:C) signaling pathways, and they detected what was interpreted as MyD88-5 in human PBMC by Western blot. However, we did not detect MyD88-5 mRNA or protein in myeloid cells from either human or mouse. Even in MyD88-5/GFP transgenic mice, where multiple copies of the transgene were expressed under MyD88-5's native promoter/enhancer, no GFP expression was detected in CD11b+ macrophages (Fig. S2). In our hands, the commercial antibody used by Carty et al. (26) detected recombinant MyD88-5, but also reacted in Western blots with a protein of similar apparent molecular weight whose abundance was unaffected by genetic ablation of MyD88-5 (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070868/DC1). Macrophages from MyD88-5 knockout mice produced similar levels of TNF and MCP-1 as their wild-type counterparts in response to various TLR ligands, including LPS, poly(I:C), Pam3Cys, and CpG. Thus, in mouse, MyD88-5 does not have a nonredundant role in regulating macrophage responses to poly(I:C) and LPS. Our anti–MyD88-5 antibody does not produce a detectable signal in Western blots of LPS-treated human monocytes (unpublished data).
The highest expression of MyD88-5 was found in brain, which is consistent with the previous clue as to MyD88-5's function in the development of olfactory neurons in the worm (18). Brain MyD88-5 was confined to neurons, in which it was localized in part to mitochondria and microtubules. Overexpression of MyD88-5 forced mitochondria to congregate at the microtubule-organizing center, raising the possibility that physiologic levels of MyD88-5 may regulate mitochondrial traffic along microtubules. This traffic may benefit from specialized molecular machinery in neurons, the cell type in which mitochondria are likely to travel longer distances than in any other type of cell (48). This interpretation is supported by the close resemblance of mitochondrial distribution in our MyD88-5–overexpressing cells and in cells in which mitochondrial traffic along microtubules was disrupted via interference with the motor protein kinesin by genetic disruption (49) or TNF treatment (50). Moreover, overexpression of proteins involved in mitochondrial fission, fusion, and trafficking decreased mitochondrial movement, promoted a rounded rather than elongated mitochondrial morphology and increased the cells' susceptibility to apoptosis (48, 51–57).
We also found that mouse MyD88-5, like its C. elegans counterpart (36), can bind JNK3. Moreover, mouse MyD88-5 recruits JNK3 to mitochondria. This is consistent with the tendency of JNK pathway core components and their regulators to form complexes that are targeted to intracellular organelles via scaffolding proteins.
Diverse stresses, such as oxidative injury and inflammatory cytokines, induce activation (58) and translocation of JNKs to mitochondria, where JNKs promote apoptosis (37, 59) by multiple mechanisms. JNK3 binds and inhibits the antiapoptotic proteins Bcl-2 (60) and Bcl-xL (37), mediates phosphorylation and oligomerization of the proapoptotic protein BAD (61), and promotes translocation of the proapoptotic protein BAX from the cytoplasm to mitochondria (62). These actions may help account for JNK3's important role in stress-induced neuronal apoptosis and in the pathogenesis of stroke and neurodegenerative diseases (43, 45). During ischemia-reperfusion injury, CA1 hippocampal neurons showed an increased level of activated JNKs and translocation of BAD to the outer mitochondrial membrane (40, 41). During transient focal cerebral ischemia, the JNK pathway mediated Bax activation and induced neuronal apoptosis (44). Conversely, overexpression of JNK-interacting protein-1 sequestered JNKs in the cytosol, conferring protection against neuronal death induced by nerve growth factor withdrawal (42). Targeted deletion of Jnk3 protected mice from seizure and from death of hippocampal neurons induced by injection of a glutamate receptor agonist (46). Cultured embryonic hippocampal neurons from jnk3-null mice retained cytochrome c inside the mitochondria after OGD, whereas wild-type cells showed diffused cytochrome c staining (43). Consistent with previous findings, MyD88-5 deficiency partially protected hippocampal neurons from OGD-induced death. It will be of interest to test if MyD88-5 helps mediate neuronal fate decisions in response to infectious agents.
This study has identified an unexpected function of mammalian MyD88-5 in regulating neuron survival, probably via association with JNK3. However, there are likely to be additional partners and functions for MyD88-5. JNK3 is not known to be expressed in T cells, and its expression in rat brain has been demonstrated only in the pyramidal cells in the hippocampus and in the cortex (63), as confirmed for pyramidal cells in human brain (64). MyD88-5 expression is broader. It is possible that MyD88-5 may contribute to immunity in T lymphocytes, which are the only cells besides neurons in which we have been able to confirm its expression.
MATERIALS AND METHODS
Mice.
C57BL/6 (B6) mice were purchased from The Jackson Laboratory. Animal studies were approved by the Institutional Animal Care and Use Committee of Weill Medical College of Cornell University.
cDNA cloning.
The full-length or different domains of MyD88-5 or Jnk3α1 cDNA were cloned using RT-PCR from RNA prepared from the brain of a C57BL/6 mouse. A mammalian expression plasmid containing the full-length human MyD88-5 cDNA (pUNO-hSARM1a) was obtained from InvivoGen. GFP or RFP fusion proteins of MyD88-5 or JNK3 were generated by inserting individual cDNAs lacking stop codons upstream of fluorescent proteins in pEGFP-N1 (Clontech) or modified pDsRed-Express1 (Clontech) with a CMV promoter.
Enrichment of mouse splenocyte subpopulations and human peripheral blood leukocytes.
T or B cells were depleted from mouse splenocytes using mouse Pan T or Pan B magnetic beads (Dynal). Human T or B cells were enriched from human peripheral blood with T or B cell negative isolation kits (Dynal). PMNs were separated from monocytes and lymphocytes by Polymorphprep (Axis-Shield PoC AS) according to the manufacturer's instruction. The monocytes and lymphocytes were then separated by incubating the cells in a plastic dish for 2 h at 37°C. Floating cells were collected as a lymphocyte-enriched population, and adherent cells were washed and collected as a monocyte-enriched population.
Measurement of mRNA expression.
The ready to use tissue mRNA membrane (BALB/c male; Stratagene) was hybridized with a cDNA fragment corresponding to the two SAM domains and part of the TIR domain of MyD88-5 (1,200–1,816 bp). Quantitative RT-PCR was performed as previously described (65). The probe and primer set sequences for mouse MyD88-5 are 5′6-FAM d(AGCGGCTCGCAATTTTGTCCTGG) BHQ-1 3′ 5′-GACAAGCTTATCCAAAGCGTCA-3′ and 5′-CGCCCCAGCAGACAGC-3′. The sequences for human MyD88-5 are 5′6-FAM d(CACCCGCAAGAGGTTCTTTAGGGAGC) BHQ-1 3′ 5′-TGGGCATGAAATCGGGC-3′ and 5′-CGAAGGTCTTGAGCTCCGTG-3′.
Generation of myd88-5/GFP BAC transgenic mice.
MyD88-5/GFP fusion transgenic mice were generated according the method of Gong et al. (66). A pair of DNA fragments (527 and 377 bp), which was homologous to the flanking sequences of a site right before and after the MyD88-5 stop codon, was inserted into a shuttle vector pLD53.SC-A-E-B (obtained from N. Heintz, The Rockefeller University, New York, NY) at the sites before and after a GFP gene. Integration of GFP into BAC was achieved by homologous recombination of the modified shuttle plasmid in a DH10 bacterial clone that harbors a BAC plasmid containing the C57BL/6J myd88-5 gene (RP23-399H5; Children's Hospital Oakland Research Institute). Modified BAC DNA was purified and injected into fertilized one-cell embryos (C57BL/6J × CBA/CA), which were then transplanted into pseudopregnant C57BL/6J female mice. Founder mice were identified by Southern blot analyses using a probe corresponding to exon 9. The injections and the transplantations were done by The Transgenic Mouse Core Facility shared by Weill Medical College and Sloan Kettering Institute.
Targeted disruption of the myd88-5 gene.
A 1,885-bp-long DNA fragment for the left arm (SA) and a 6,880-bp-long DNA fragment for the right arm (LA) were PCR amplified from 129/SvJ mouse genomic DNA with an expanded 20 kb plus PCR system (Roche). The two arms were cloned with a PGK-neomycin cassette (67) and a diphtheria toxin gene was added for negative selection (68). The resulting construct was linearized at a unique XhoI enzyme site before transfection into 129/SvJ ES cells. 3 out of 192 clones selected with G418 were identified as positive for homologous recombination and used for injection into C57BL/6 blastocysts. Chimeras were identified and crossed with C57BL/6J for germline transmission. Genomic DNA prepared from tails was digested with SphI restriction enzyme for Southern analysis with a radiolabeled probe corresponding to exon 2. Transfection, selection, and screening of ES cells were done in the Gene Targeting Facility at The Rockefeller University. Pups with germline transmission were intercrossed at Weill Medical College to generate mice with wild-type, heterozygous, or homozygous knockout alleles.
Generation of antibody to MyD88-5.
A full-length MyD88-5 cDNA was subcloned into Novagen's pET30b(+) for a fusion protein with hexahistidine attached to its N terminus. BL21 (DE3) cells (2 liters) were transformed with this plasmid and grown until the OD600 reached 0.4. Protein expression was induced for 3 h with 1 mM isopropyl-(-D-thiogalactopyranoside), at which time most of the recombinant MyD88-5 was found in inclusion bodies. The bacterial pellet was lysed at room temperature for 20 min with 100 ml of Tris buffer (50 mM, pH 8.0) containing 100 mM NaCl, 1 mM EDTA, and 1 mg/ml lysozyme, and centrifuged (500 g, 10 min). The pellets were purified further by resuspending in 50 ml of the same buffer with 0.1% deoxycholate sodium salt. After 10 min of incubation on ice, 8 mM MgCl2 and 10 μg/ml DNase I were added to the samples for an additional 1 h. Samples were centrifuged again at 10,000 g for 10 min at 4°C. The pellet was first washed with 50 ml of Tris buffer (50 mM, pH 8.0) containing 100 mM NaCl, 1 mM EDTA, and 1% NP-40, and then again with 50 ml of Tris buffer (50 mM, pH 8.0) with 1 M urea. The pellet was dissolved in the sample buffer and run on a 9% SDS-PAGE gel. The protein band corresponding to MyD88-5, which was visualized by Coomassie staining, was cut out and the recombinant protein was eluted by a previously described method (69). Recombinant MyD88-5 was concentrated with a Centricon column (10 kD cut-off; Millipore) and used for immunization of chickens to generate anti–mouse MyD88-5 fowl immunoglobulin (IgY).
Antibody was affinity purified by incubating 2 ml of purified IgY (27 mg/ml) with a nitrocellulose membrane (10 cm2) on which 2 mg of purified recombinant MyD88-5 had been blotted. After overnight incubation at 4°C, the membrane was washed two times each with TBS and PBS for 5 min. Membrane-bound antibody was eluted twice with 100 mM glycine buffer, pH 2.5, for 10 min. Eluted antibody was neutralized with 1 M Tris buffer, pH 8.5, and stored in the presence of 5 mM sodium azide and 1 mg/ml BSA.
Transfection, immunoprecipitation, and immunoblot.
COS-1 cells cultured in 10-cm dishes at 80% confluency were transfected for 18 h with 3 μg of expression plasmids pJNK3/EGFP or pEGFP together with 3 μg of pMyD88-5 with Lipofectamine 2000 (Sigma-Aldrich). Hippocampi were dissected from 8-wk-old C57BL/6 mice and coronal slices (thickness 150 μM) were cut. Slices were transferred to Millicell CM membrane inserts (30 MM, 0.4 μm; Millipore) and the inserts were placed into 6-well plates containing 0.5 ml culture medium (25% horse serum, 50% Eagle's basal medium, 25% HBSS, 5 mg/ml glucose) in each well. Plates were exposed to ultraviolet light in a cell culture hood for 1 min (∼120 Joules/m2). Hippocampal and cell lysates were prepared in 0.5 ml IP buffer (PBS supplemented with 5 mM EDTA, 0.5% Triton X-100, and protease inhibitors) and precleared with protein G beads (4°C for 1 h) for immunoprecipitation or boiled directly in Laemmli sample buffer for immunoblot. Precleared lysates were incubated for 4 h with anti-GFP (BD Bioscience) or anti-JNK1,3 (eBioscience) mixed with Protein G Sepharose (GE Healthcare) at 4°C. Washed beads were boiled in Laemmli buffer. Proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane for immunoblotting with chicken anti–MyD88-5, mouse anti-GFP (Berkeley), or rabbit anti-JNK3 (Millipore), and they were visualized with an enhanced chemiluminescence system (Millipore).
Epifluorescent and confocal microscopy.
COS-1 cells grown on coverslips in 12-well plates were transfected for 18 h with 400 ng of plasmids mixed with 0.8 μl of Lipofectamine 2000 (Invitrogen). For mitochondrial detection, cells were stained with 10 nM MitoTracker Red CMXRos (Invitrogen) for 20 min before fixation. For microtubule and Golgi detection, cells were fixed with ice-cold methanol for 10 min or 4% paraformaldehyde in PBS for 5 min at room temperature, followed by permeabilization with saponin (0.0375%). Fixed cells were stained with anti-tubulin (1:200; GE Healthcare) or anti–γ-adaptin antibody (1:100; BD Biosciences), followed by Cy3-conjugated Fab fragment of donkey anti–mouse IgG (1:800; Jackson ImmunoResearch Laboratories) for detection of microtubules or Golgi, respectively. Nuclei were stained with 10 ng/ml DAPI.
Epifluorescent images were acquired using a digital camera (Coolpix 5000; Nikon) attached to a fluorescent microscope (Bx60; Olympus) with a 1,000× magnification lens and a 3× zoom lens on the camera. Confocal images were taken with a spectral confocal system (TCS SP2; Leica). The acoustooptical beam-splitter system and sequential image recording were used for image collection. The excitation intensities and spectral detection windows were systematically adjusted to optimize signals and avoid cross talk between channels. Single-channel excitation and dual-channel recording before data collection routinely confirmed the absence of cross talk between the channels.
Isolation of mitochondria and microtubules.
Mitochondria were isolated from mouse brain using a method modified from Kang et al. (70). In brief, 2 mouse brains were washed, minced in 5 ml of ice-cold Mito buffer (10 mM Tris HCl, pH 7.8, plus 0.2 mM EDTA, 0.25 M sucrose, and protease inhibitors), and homogenized. Supernatant after centrifugation (800 g, 10 min at 4°C) was collected and centrifuged again at 10,000 g at 4°C for 10 min to enrich the mitochondrial fraction. The resulting pellet was resuspended in 1 ml of the same buffer and spun at 300 g at 4°C for 5 min to remove bigger organelles. The supernatant was then collected and centrifuged at 10,000 g at 4°C for 10 min to pellet the mitochondrial fraction. The pellet was washed once and resuspended with Mito buffer for Western blot analysis.
Microtubule-associated proteins were isolated according to the method of Vallee et al. (71). Brains from 10 mice were pooled and homogenized in 7.5 ml of 0.1 M PEM buffer (0.1 M Pepes, pH 6.9, with 1 mM EGTA and 0.5 mM MgCl2) supplemented with protease inhibitors at 4°C. After centrifugation at 100,000 g for 60 min at 4°C, supernatants were then divided into two aliquots. One aliquot was incubated at 37°C for 15 min with 20 μM of Taxol and 1 mM of GTP to polymerize microtubules and the other aliquot was kept at 4°C with addition of 0.1 mM colchicine (negative control). Polymerized microtubules were isolated by centrifugation on a sucrose gradient (10% sucrose, 30,000 g, 30 min at 37°C). The pellet containing microtubules and microtubule-binding proteins was rinsed with warm PEM buffer and subjected to Western blot analysis.
Organotypic mouse hippocampal slice culture and OGD.
Hippocampi were dissected aseptically from 5–6-d-old myd88-5 −/− mice and their wild-type or hemizygous littermates, and 350-μM-thick coronal slices were cut using a McIlwain tissue chopper (Vibratome Company). Slices were transferred to Millicell CM membrane inserts (30 MM, 0.4 μm; Millipore), and the inserts were placed into 6-well plates containing a 1-ml-slice of culture medium (25% horse serum, 50% Eagle's basal medium, 25% HBSS, 5 mg/ml glucose) in each well. The cultures were maintained in a humidified chamber at 37°C with 5% CO2 for 13 d, and the medium was changed twice a week. On day 13, slides were incubated in fresh medium containing the cell death marker PI (2.5 μg/ml; Sigma-Aldrich). 1 d later, baseline fluorescence images (Fbasal) were taken using a fluorescence microscope (Eclipse; Nikon) equipped with a digital camera (Retiga Exi; QImaging) connected to a computer (Macintosh) running IPLab software (Scanalytics, Inc.). Procedures for OGD and assessment of neuronal injury were as follows. Slices were placed in OGD buffer (125 mM NaCl, 5 mM KCl, 1.2 mM Na2HPO4, 26 mM NaHCO3, 1.8 mM CaCl2, 0.9 mM MgCl2, and 10 mM Hepes, pH 7.4), transferred to an OGD chamber (Billups-Rothenberg), and flushed (21 liters/min for 5 min) with a gas mixture containing 95% N2 and 5% CO2. The chamber was sealed and placed into an incubator at 37°C for 1 h. Control slices were rinsed with OGD buffer containing 10 mM glucose and incubated in normoxic conditions. After OGD, the slices were transferred to normal culture medium containing PI and incubated for 24 h before fluorescence images were taken (FOGD). The slices were treated with 100 μM NMDA and PI for an additional 24 h, and images were captured for assessment of maximal fluorescence intensity (Fmax). Slices were imaged in the same order and orientation. The camera exposure settings were carefully adjusted so that Fbasal was detectable and Fmax did not saturate the camera. The same settings (exposure time, filter setting, and gain) were used throughout the experiments. The percentage of cell death in CA1 was calculated with the formula (FOGD − Fbasal)/(Fmax − Fbasal) × 100. Two-group comparisons were analyzed by the two-tailed Student's t test for independent samples. P values <0.05 were considered statistically significant.
Online supplemental material.
Fig. S1 illustrates the domain organization of MyD88-5 and its position in a phylogenic tree within the TIR domain superfamily. Fig. S2 indicates that GFP could not be detected by flow cytometry in thymocytes or CD11b ± splenocytes (macrophages) of MyD88-5/GFP BAC transgenic mice. Fig. S3 shows that ablation of MyD88-5 has no effect on macrophage responses to various TLR ligands. Fig. S4 shows that a commercial anti-SARM (MyD88-5) antibody recognizes a protein in organs from MyD88-5 knockout mice.The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070868/DC1.
Supplemental Material
Acknowledgments
We thank Krista LaPerle for pathological evaluation of the MyD88-5 knockout mice; Lee Gould for taking confocal images of the transgenic mouse brain; Nat Heinz for granting the usage of the BAC transgenic shuttle vector; Chinghwa Sung for scientific discussion; and Nichole Chan for the assistance with mouse maintenance and genotyping.
This work was supported by National Institutes of Health grants AI30165/GM61710 (A. Ding) and NS34179/NS35806 (C. Iadecola) and the Dana Foundation. The Department of Microbiology and Immunology thanks the William Randolph Hearst Foundation.
The authors have no conflicting financial interests.
Abbreviations used: BAC, bacterial artificial chromosome; MAL, MyD88 adaptor–like; OGD, oxygen and glucose deprivation; PI, propidium iodide; RFP, red fluorescent protein; SAM, sterile α motif ; SARM, sterile α and HEAT/Armadillo motifs containing protein; TICAM, TIR domain–containing adaptor molecule; TIR, Toll-interleukin 1 receptor; TIRAP, TIR domain–containing adaptor protein; TIRP, TIR domain–containing protein; TLR, Toll-like receptor; TRAM, TRIF-related adaptor molecule; TRIF, TIR-domain containing adaptor inducing interferon-β.
C. Nathan and A. Ding contributed equally to this paper.
References
- 1.Alexopoulou, L., A.C. Holt, R. Medzhitov, and R.A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. [DOI] [PubMed] [Google Scholar]
- 2.Xu, Y., X. Tao, B. Shen, T. Horng, R. Medzhitov, J.L. Manley, and L. Tong. 2000. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 408:111–115. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgerald, K.A., E.M. Palsson-McDermott, A.G. Bowie, C.A. Jefferies, A.S. Mansell, G. Brady, E. Brint, A. Dunne, P. Gray, M.T. Harte, et al. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 413:78–83. [DOI] [PubMed] [Google Scholar]
- 4.Horng, T., G.M. Barton, and R. Medzhitov. 2001. TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol. 2:835–841. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto, M., S. Sato, H. Hemmi, H. Sanjo, S. Uematsu, T. Kaisho, K. Hoshino, O. Takeuchi, M. Kobayashi, T. Fujita, et al. 2002. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 420:324–329. [DOI] [PubMed] [Google Scholar]
- 6.Hoebe, K., X. Du, P. Georgel, E. Janssen, K. Tabeta, S.O. Kim, J. Goode, P. Lin, N. Mann, S. Mudd, et al. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 424:743–748. [DOI] [PubMed] [Google Scholar]
- 7.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161–167. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto, M., S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Sanjo, O. Takeuchi, M. Sugiyama, M. Okabe, K. Takeda, and S. Akira. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. [DOI] [PubMed] [Google Scholar]
- 9.Bin, L.H., L.G. Xu, and H.B. Shu. 2003. TIRP, a novel Toll/interleukin-1 receptor (TIR) domain-containing adapter protein involved in TIR signaling. J. Biol. Chem. 278:24526–24532. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K.A., D.C. Rowe, B.J. Barnes, D.R. Caffrey, A. Visintin, E. Latz, B. Monks, P.M. Pitha, and D.T. Golenbock. 2003. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the toll adapters TRAM and TRIF. J. Exp. Med. 198:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto, M., S. Sato, H. Hemmi, S. Uematsu, K. Hoshino, T. Kaisho, O. Takeuchi, K. Takeda, and S. Akira. 2003. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4:1144–1150. [DOI] [PubMed] [Google Scholar]
- 12.Mink, M., B. Fogelgren, K. Olszewski, P. Maroy, and K. Csiszar. 2001. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 74:234–244. [DOI] [PubMed] [Google Scholar]
- 13.Andrade, M.A., C. Perez-Iratxeta, and C.P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117–131. [DOI] [PubMed] [Google Scholar]
- 14.Aviv, T., Z. Lin, G. Ben-Ari, C.A. Smibert, and F. Sicheri. 2006. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 13:168–176. [DOI] [PubMed] [Google Scholar]
- 15.Coates, J.C. 2003. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13:463–471. [DOI] [PubMed] [Google Scholar]
- 16.Couillault, C., N. Pujol, J. Reboul, L. Sabatier, J.F. Guichou, Y. Kohara, and J.J. Ewbank. 2004. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5:488–494. [DOI] [PubMed] [Google Scholar]
- 17.Liberati, N.T., K.A. Fitzgerald, D.H. Kim, R. Feinbaum, D.T. Golenbock, and F.M. Ausubel. 2004. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA. 101:6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang, C.F., and C.I. Bargmann. 2005. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 19:270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang, S., R.J. Johnston Jr., and O. Hobert. 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 17:2123–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troemel, E.R., A. Sagasti, and C.I. Bargmann. 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 99:387–398. [DOI] [PubMed] [Google Scholar]
- 21.Wes, P.D., and C.I. Bargmann. 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 410:698–701. [DOI] [PubMed] [Google Scholar]
- 22.Sagasti, A., N. Hisamoto, J. Hyodo, M. Tanaka-Hino, K. Matsumoto, and C.I. Bargmann. 2001. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 105:221–232. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 11:115–122. [DOI] [PubMed] [Google Scholar]
- 24.Horng, T., G.M. Barton, R.A. Flavell, and R. Medzhitov. 2002. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 420:329–333. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser, W.J., and M.K. Offermann. 2005. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J. Immunol. 174:4942–4952. [DOI] [PubMed] [Google Scholar]
- 26.Carty, M., R. Goodbody, M. Schroder, J. Stack, P.N. Moynagh, and A.G. Bowie. 2006. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 7:1074–1081. [DOI] [PubMed] [Google Scholar]
- 27.Bonnert, T.P., K.E. Garka, P. Parnet, G. Sonoda, J.R. Testa, and J.E. Sims. 1997. The cloning and characterization of human MyD88: a member of an IL-1 receptor related family. FEBS Lett. 402:81–84. [DOI] [PubMed] [Google Scholar]
- 28.Hardiman, G., N.A. Jenkins, N.G. Copeland, D.J. Gilbert, D.K. Garcia, S.L. Naylor, R.A. Kastelein, and J.F. Bazan. 1997. Genetic structure and chromosomal mapping of MyD88. Genomics. 45:332–339. [DOI] [PubMed] [Google Scholar]
- 29.Lord, K.A., B. Hoffman-Liebermann, and D.A. Liebermann. 1990. Nucleotide sequence and expression of a cDNA encoding MyD88, a novel myeloid differentiation primary response gene induced by IL6. Oncogene. 5:1095–1097. [PubMed] [Google Scholar]
- 30.Seki, E., H. Tsutsui, H. Nakano, N. Tsuji, K. Hoshino, O. Adachi, K. Adachi, S. Futatsugi, K. Kuida, O. Takeuchi, et al. 2001. Lipopolysaccharide-induced IL-18 secretion from murine Kupffer cells independently of myeloid differentiation factor 88 that is critically involved in induction of production of IL-12 and IL-1beta. J. Immunol. 166:2651–2657. [DOI] [PubMed] [Google Scholar]
- 31.Raha, D., Q.D. Nguyen, and A. Garen. 1990. Molecular and developmental analyses of the protein encoded by the Drosophila gene ectodermal. Dev. Genet. 11:310–317. [DOI] [PubMed] [Google Scholar]
- 32.Mody, M., Y. Cao, Z. Cui, K.Y. Tay, A. Shyong, E. Shimizu, K. Pham, P. Schultz, D. Welsh, and J.Z. Tsien. 2001. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc. Natl. Acad. Sci. USA. 98:8862–8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong, S., C. Zheng, M.L. Doughty, K. Losos, N. Didkovsky, U.B. Schambra, N.J. Nowak, A. Joyner, G. Leblanc, M.E. Hatten, and N. Heintz. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 425:917–925. [DOI] [PubMed] [Google Scholar]
- 34.Wanka, F., and E.J. Van Zoelen. 2003. Cellular organelle transport and positioning by plasma streaming. Cell. Mol. Biol. Lett. 8:1035–1045. [PubMed] [Google Scholar]
- 35.Costanzo, M.C., J.D. Hogan, M.E. Cusick, B.P. Davis, A.M. Fancher, P.E. Hodges, P. Kondu, C. Lengieza, J.E. Lew-Smith, C. Lingner, et al. 2000. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 28:73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, S., C.M. Armstrong, N. Bertin, H. Ge, S. Milstein, M. Boxem, P.O. Vidalain, J.D. Han, A. Chesneau, T. Hao, et al. 2004. A map of the interactome network of the metazoan C. elegans. Science. 303:540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kharbanda, S., S. Saxena, K. Yoshida, P. Pandey, M. Kaneki, Q. Wang, K. Cheng, Y.N. Chen, A. Campbell, T. Sudha, et al. 2000. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J. Biol. Chem. 275:322–327. [DOI] [PubMed] [Google Scholar]
- 38.Bowman, A.B., A. Kamal, B.W. Ritchings, A.V. Philp, M. McGrail, J.G. Gindhart, and L.S. Goldstein. 2000. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 103:583–594. [DOI] [PubMed] [Google Scholar]
- 39.Verhey, K.J., D. Meyer, R. Deehan, J. Blenis, B.J. Schnapp, T.A. Rapoport, and B. Margolis. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152:959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carboni, S., B. Antonsson, P. Gaillard, J.P. Gotteland, J.Y. Gillon, and P.A. Vitte. 2005. Control of death receptor and mitochondrial-dependent apoptosis by c-Jun N-terminal kinase in hippocampal CA1 neurones following global transient ischaemia. J. Neurochem. 92:1054–1060. [DOI] [PubMed] [Google Scholar]
- 41.Dluzniewska, J., M. Beresewicz, U. Wojewodzka, B. Gajkowska, and B. Zablocka. 2005. Transient cerebral ischemia induces delayed proapoptotic Bad translocation to mitochondria in CA1 sector of hippocampus. Brain Res. Mol. Brain Res. 133:274–280. [DOI] [PubMed] [Google Scholar]
- 42.Harding, T.C., L. Xue, A. Bienemann, D. Haywood, M. Dickens, A.M. Tolkovsky, and J.B. Uney. 2001. Inhibition of JNK by overexpression of the JNL binding domain of JIP-1 prevents apoptosis in sympathetic neurons. J. Biol. Chem. 276:4531–4534. [DOI] [PubMed] [Google Scholar]
- 43.Kuan, C.Y., A.J. Whitmarsh, D.D. Yang, G. Liao, A.J. Schloemer, C. Dong, J. Bao, K.J. Banasiak, G.G. Haddad, R.A. Flavell, et al. 2003. A critical role of neural-specific JNK3 for ischemic apoptosis. Proc. Natl. Acad. Sci. USA. 100:15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuno, S., A. Saito, T. Hayashi, and P.H. Chan. 2004. The c-Jun N-terminal protein kinase signaling pathway mediates Bax activation and subsequent neuronal apoptosis through interaction with Bim after transient focal cerebral ischemia. J. Neurosci. 24:7879–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resnick, L., and M. Fennell. 2004. Targeting JNK3 for the treatment of neurodegenerative disorders. Drug Discov. Today. 9:932–939. [DOI] [PubMed] [Google Scholar]
- 46.Yang, D.D., C.Y. Kuan, A.J. Whitmarsh, M. Rincon, T.S. Zheng, R.J. Davis, P. Rakic, and R.A. Flavell. 1997. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 389:865–870. [DOI] [PubMed] [Google Scholar]
- 47.Kawano, T., J. Anrather, P. Zhou, L. Park, G. Wang, K.A. Frys, A. Kunz, S. Cho, M. Orio, and C. Iadecola. 2006. Prostaglandin E2 EP1 receptors: downstream effectors of COX-2 neurotoxicity. Nat. Med. 12:225–229. [DOI] [PubMed] [Google Scholar]
- 48.Chan, D.C. 2006. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 125:1241–1252. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, Y., Y. Kanai, Y. Okada, S. Nonaka, S. Takeda, A. Harada, and N. Hirokawa. 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 93:1147–1158. [DOI] [PubMed] [Google Scholar]
- 50.De Vos, K., F. Severin, F. Van Herreweghe, K. Vancompernolle, V. Goossens, A. Hyman, and J. Grooten. 2000. Tumor necrosis factor induces hyperphosphorylation of kinesin light chain and inhibits kinesin-mediated transport of mitochondria. J. Cell Biol. 149:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breckenridge, D.G., M. Stojanovic, R.C. Marcellus, and G.C. Shore. 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ebneth, A., R. Godemann, K. Stamer, S. Illenberger, B. Trinczek, and E. Mandelkow. 1998. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J. Cell Biol. 143:777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jagasia, R., P. Grote, B. Westermann, and B. Conradt. 2005. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 433:754–760. [DOI] [PubMed] [Google Scholar]
- 54.Karbowski, M., and R.J. Youle. 2003. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 10:870–880. [DOI] [PubMed] [Google Scholar]
- 55.Matarrese, P., A. Tinari, E. Mormone, G.A. Bianco, M.A. Toscano, B. Ascione, G.A. Rabinovich, and W. Malorni. 2005. Galectin-1 sensitizes resting human T lymphocytes to Fas (CD95)-mediated cell death via mitochondrial hyperpolarization, budding, and fission. J. Biol. Chem. 280:6969–6985. [DOI] [PubMed] [Google Scholar]
- 56.Santel, A., S. Frank, B. Gaume, M. Herrler, R.J. Youle, and M.T. Fuller. 2003. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 116:2763–2774. [DOI] [PubMed] [Google Scholar]
- 57.Sugioka, R., S. Shimizu, and Y. Tsujimoto. 2004. Fzo1, a protein involved in mitochondrial fusion, inhibits apoptosis. J. Biol. Chem. 279:52726–52734. [DOI] [PubMed] [Google Scholar]
- 58.Kyriakis, J.M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807–869. [DOI] [PubMed] [Google Scholar]
- 59.Ito, Y., N.C. Mishra, K. Yoshida, S. Kharbanda, S. Saxena, and D. Kufe. 2001. Mitochondrial targeting of JNK/SAPK in the phorbol ester response of myeloid leukemia cells. Cell Death Differ. 8:794–800. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto, K., H. Ichijo, and S.J. Korsmeyer. 1999. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol. Cell. Biol. 19:8469–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhakar, A.L., J.L. Howell, C.E. Paul, A.H. Salehi, E.B. Becker, F. Said, A. Bonni, and P.A. Barker. 2003. Apoptosis induced by p75NTR overexpression requires Jun kinase-dependent phosphorylation of Bad. J. Neurosci. 23:11373–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuruta, F., J. Sunayama, Y. Mori, S. Hattori, S. Shimizu, Y. Tsujimoto, K. Yoshioka, N. Masuyama, and Y. Gotoh. 2004. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 23:1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohit, A.A., J.H. Martin, and C.A. Miller. 1995. p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron. 14:67–78. [DOI] [PubMed] [Google Scholar]
- 64.Bingham, J., and S. Sudarsanam. 2000. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 16:660–661. [DOI] [PubMed] [Google Scholar]
- 65.Shi, S., C. Nathan, D. Schnappinger, J. Drenkow, M. Fuortes, E. Block, A. Ding, T.R. Gingeras, G. Schoolnik, S. Akira, et al. 2003. MyD88 primes macrophages for full-scale activation by interferon-γ yet mediates few responses to Mycobacterium tuberculosis. J. Exp. Med. 198:987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gong, S., X.W. Yang, C. Li, and N. Heintz. 2002. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 12:1992–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanks, M., W. Wurst, L. Anson-Cartwright, A.B. Auerbach, and A.L. Joyner. 1995. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 269:679–682. [DOI] [PubMed] [Google Scholar]
- 68.Yagi, T., Y. Ikawa, K. Yoshida, Y. Shigetani, N. Takeda, I. Mabuchi, T. Yamamoto, and S. Aizawa. 1990. Homologous recombination at c-fyn locus of mouse embryonic stem cells with use of diphtheria toxin A-fragment gene in negative selection. Proc. Natl. Acad. Sci. USA. 87:9918–9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Retamal, C.A., P. Thiebaut, and E.W. Alves. 1999. Protein purification from polyacrylamide gels by sonication extraction. Anal. Biochem. 268:15–20. [DOI] [PubMed] [Google Scholar]
- 70.Kang, D., J. Nishida, A. Iyama, Y. Nakabeppu, M. Furuichi, T. Fujiwara, M. Sekiguchi, and K. Takeshige. 1995. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem. 270:14659–14665. [DOI] [PubMed] [Google Scholar]
- 71.Vallee, R.B. 1982. A taxol-dependent procedure for the isolation of microtubules and microtubule-associated proteins (MAPs). J. Cell Biol. 92:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.