Abstract
Recently, a new developmental pathway for CD4 T cells that is mediated by major histocompatibility complex class II–positive thymocytes was identified (Choi, E.Y., K.C. Jung, H.J. Park, D.H. Chung, J.S. Song, S.D. Yang, E. Simpson, and S.H. Park. 2005. Immunity. 23:387–396; Li, W., M.G. Kim, T.S. Gourley, B.P. McCarthy, D.B. Sant'angelo, and C.H. Chang. 2005. Immunity. 23:375–386). We demonstrate that thymocyte-selected CD4 (T-CD4) T cells can rapidly produce interferon γ and interleukin (IL) 4 upon in vivo and in vitro T cell receptor stimulation. These T-CD4 T cells appear to be effector cells producing both T helper type 1 (Th1) and Th2 cytokines, and they maintain a potential to produce Th2 cytokines under Th1-skewing conditions in a signal transducer and activator of transcription 6–independent manner. The IL-4 mRNA level is high in CD4 single-positive thymocytes if they are selected on thymocytes, which is at least partly caused by enhanced histone acetylation of the IL-4 locus. However, mice that can generate T-CD4 T cells showed attenuated immune responses in an allergen-induced airway inflammation model, suggesting a protective role for T-CD4 T cells during an airway challenge. Our results imply that this thymic selection pathway plays an important role in determining the effector function of the resulting CD4 cells and in regulating immune response.
Upon antigen stimulation, naive CD4 T cells can differentiate into Th1, Th2, or the newly characterized Th17 effector cells, which rapidly produce IFN-γ, IL-4, or IL-17, respectively (1–4). The hallmark cytokine of Th1 cells is IFN-γ, which is instrumental for cell-mediated immunity. Th2 cells produce the prototypical cytokine IL-4, as well as IL-5 and IL-13, in controlling humoral immune responses. IL-17, collectively with other cytokines and chemokines released by activated Th17 cells, plays an important role in several inflammatory autoimmune diseases (5–10). Thus, proper regulation of Th differentiation is critical for controlling both cellular and humoral immune responses and for maintaining immune homeostasis.
The cytokines made by pathogen-activated cells of the innate immune system during T cell priming are key factors in promoting Th differentiation. It is known that type I or type II IFNs from activated NK cells or plasmacytoid DCs, collectively with IL-12 produced by APCs, direct CD4 T cells into the Th1 cell lineage. IL-4 present in the priming environment preferentially induces Th2 cell differentiation. TGF-β and IL-6 promote the development of Th17 cells that expand in response to IL-23 (11–13). The Th1 and Th2 cytokines IFN-γ and IL-4, respectively, can inhibit the production of the opposing types of cytokines by CD4 T cells and antagonize Th17 cell differentiation. Th cell fate has also been shown to be influenced by several other factors including TCR affinity, antigen dosage, co-stimulatory molecules, and the type of APC (14–17). However, it is not known whether T cell selection in the thymus affects Th cell fate in the periphery.
Recently, we and others have demonstrated that MHC class II–expressing thymocytes can efficiently mediate the positive selection of CD4 T cells in mice (18, 19). Human thymocytes express MHC class II, and thymocyte-mediated CD4 T cell selection provides a mechanism for several documented observations that could not be otherwise explained (20–28). Therefore, in humans, CD4 T cells can be selected by two pathways, and the two CD4 T cell populations likely coexist in the periphery. Because mouse thymocytes do not express MHC class II, thymocyte-mediated selection has not been considered, and the role of this pathway in Th cell fate has been neglected.
We report in this study that thymocyte-selected CD4 (T-CD4) T cells differ from thymic epithelial cell (TEC)–selected CD4 (E-CD4) T cells in that they can rapidly produce both IL-4 and IFN-γ upon in vivo as well as in vitro TCR stimulation. When differentiated into effector cells under nonpolarizing conditions, these T-CD4 T cells appear to be Th0 effector cells able to produce both Th1 and Th2 cytokines. Furthermore, they maintained the capability to produce the opposite type of cytokines when differentiated into Th1 or Th2 cell lineages.
T-CD4 cells made IL-4 shortly after stimulation and also produced Th2 cytokines under Th1-inducing conditions in a Stat6-independent manner. A high level of preformed IL-4 mRNA were detected in CD4 single-positive (SP) thymocytes if they are selected on thymocytes, which is at least partly caused by enhanced histone acetylation of the IL-4 locus. Finally, T-CD4 T cells seem to have a protective role during an airway challenge, suggesting a unique regulatory function of T-CD4 T cells during an immune response.
RESULTS
T-CD4 T cells rapidly produce both IL-4 and IFN-γ
We generated mice that express MHC class II in thymocytes (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20070321/DC1) and peripheral T cells by introducing the MHC class II transactivator (CIITA) as a transgene under control of the CD4 promoter (CIITATg) (29). CIITA is a transcription factor that is both necessary and sufficient for the expression of molecules that participate in MHC class II–restricted antigen presentation (30). Using these mice, we previously demonstrated that CD4 T cells can be positively selected via both cortical TECs (cTECs) and thymocytes (19). Therefore, CIITATg mice generate a mixed population of CD4 T cells that are selected on TECs or thymocytes, whereas the control (WT) mice have only TEC-selected CD4 T cells (19). To distinguish between the two CD4 T cell populations present in CIITATg mice, we named them E- and T-CD4 T cells to reflect the cell type mediating selection (epithelial cell– and thymocyte-selected CD4 T cells, respectively).
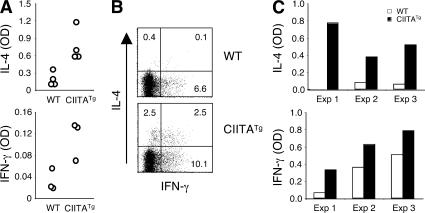
To study whether CD4 T cell function is different depending on their selection pathway, we initially examined the cytokine production potential of T-CD4 T cells upon in vivo stimulation. Although we have reported that CD4 T cells prepared from CIITATg mice produced elevated IL-4, the cytokine production upon in vivo stimulation was not studied (29). WT and CIITATg mice were injected with an anti-CD3 antibody i.v., and the levels of serum IL-4 and IFN-γ were measured 2 h later. CIITATg mice that have T-CD4 T cells showed a dramatic increase in both cytokines, suggesting an effector phenotype of T-CD4 cells (Fig. 1 A). Consistent with the in vivo induction of IL-4 and IFN-γ production, splenic CD4 T cells from CIITATg mice expressed more IFN-γ and IL-4 than WT mice 5 h after stimulation in vitro (Fig. 1 B) (29). Moreover, splenic CD4 T cells from CIITATg mice had a greater potential to produce IL-4 and IFN-γ than WT mice after a 2-d stimulation in vitro (Fig. 1 C).
Figure 1.
T-CD4 T cells rapidly produce high levels of IL-4 and IFN-γ. (A) 2 h after tail vein injection of PBS or 10 μg anti-CD3 antibody, WT and CIITATg mice were killed and blood was collected. Cytokine levels in the serum were determined by ELISA. Each symbol in the histograms represents an individual mouse. (B) Splenocytes from the indicated mice were stimulated with PMA and ionomycin for 5 h, followed by ICS. Plots were gated on NK1.1− CD4 T cells. The percentage of positive cells in each quadrant is shown. (C) Splenic CD4 T cells from WT and CIITATg mice were stimulated with anti-CD3 and anti-CD28 for 2 d. Cytokines in the culture supernatants were determined by ELISA. Results from three independent experiments are shown.
T-CD4 Th1 cells produce Th2 cytokines in addition to IFN-γ
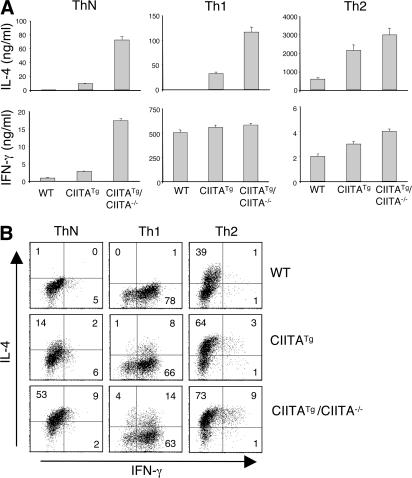
Next, we investigated the effector function of T-CD4 T cells in comparison to E-CD4 T cells. For this set of experiments, we included CIITATg/CIITA−/− mice that only generate T-CD4 T cells because of a deficiency of MHC class II expression in TECs (19). Because there are more peripheral CD4 T cells of the effector/memory type (CD44hiCD45RBlo) in CIITATg and CIITATg/CIITA−/− mice than in WT mice (i.e., 42 ± 9%, 41 ± 7%, and 27 ± 6% among splenic CD4 T cells, respectively; Fig. S1 B), we sorted naive CD4 T cells (CD44loCD45RBhi) from total peripheral CD4 T cells (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070321/DC1). Sorted naive cells were then differentiated under neutral (ThN)-, Th1-, or Th2-inducing conditions. ThN cells from WT mice produced a small amount of IFN-γ and almost undetectable levels of IL-4 (Fig. 2, A and B). In contrast, CIITATg cells made a lot more IFN-γ and IL-4 than WT cells. Moreover, CIITATg/CIITA−/− cells produced very high levels of both IFN-γ and IL-4 (Fig. 2, A and B). Th1 cells from WT mice predominantly made IFN-γ but not IL-4, whereas both CIITATg and CIITATg/CIITA−/− Th1 cells produced IL-4 in addition to IFN-γ (Fig. 2, A and B) (29). The amount of IL-4 was much greater in CIITATg/CIITA−/− than in CIITATg Th1 cells, whereas IFN-γ levels were comparable among the three. When we examined Th2 cells, both CIITATg and CIITATg/CIITA−/− Th2 cells produced more IL-4 than WT cells. Unlike Th1 cells, Th2 cells from CIITATg or CIITATg/CIITA−/− mice produced a moderately elevated level of IFN-γ. The intracellular cytokine stating (ICS) data supported the dual cytokine production by CIITATg and CIITATg/CIITA−/− cells with generally more cytokine-producing CIITATg/CIITA−/− cells (Fig. 2 B). Total CD4 T cells from the same mice showed a similar cytokine pattern as naive CD4 T cells (Fig. S3). Other Th2 cytokines, such as IL-5 and IL-13, were also increased in CIITATg or CIITATg/CIITA−/− CD4 T cells (unpublished data) (29). Because CIITATg and CIITATg/CIITA−/− CD4 T cells can express the opposite cytokines, even under Th1- or Th2-skewing conditions, and the Th1 cells expressed a relatively more prominent amount of IL-4, we subsequently focused on the characterization of Th2 cytokine production by T-CD4 Th1 cells.
Figure 2.
Th1 cells from T-CD4 T cells produce the Th2 cytokine IL-4 as well as IFN-γ. Sorted naive (CD45RBhiCD44lo) CD4 T cells from WT, CIITATg, and CIITATg/CIITA−/− mice were cultured under neutral, Th1-, or Th2-skewing conditions for 6 d, as described in Materials and methods. Differentiated cells were subsequently restimulated with plate-coated anti-CD3 overnight, and culture supernatants were collected and analyzed for IFN-γ and IL-4 production by ELISA (A). The error bars represent the mean ± SD. For intracellular cytokine analysis (B), differentiated neutral, Th1, or Th2 cells were restimulated with PMA plus ionomycin for 5 h, as described in Materials and methods. After fixation and permeabilization, the cells were stained with PE-conjugated anti–IFN-γ and allophycocyanin-conjugated anti–IL-4 and analyzed by FACS. Numbers in the dot plots represent the percentages of cytokine-positive CD4 T cells. All experiments were repeated at least twice.
The thymocyte-mediated T cell selection pathway is responsible for the phenotype of T-CD4 T cells
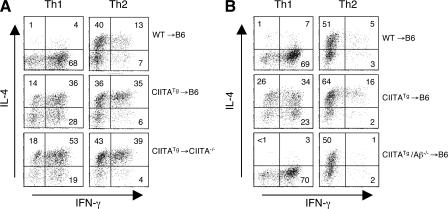
To further substantiate the role of the CD4 T cell selection pathway in cytokine production potential, we used BM chimeric mice. We previously showed that in the WT hosts (C57BL/6), CIITATg CD4 cells are developed on thymocytes as well as on cTECs, whereas in MHC class II–deficient hosts, CD4 cells are only selected on thymocytes (19). Using this principle, we generated and examined three types of chimeric mice: WT→B6, CIITATg→B6, and CIITATg→CIITA−/− mice, which generate E-CD4, E- and T-CD4, and T-CD4 T cells, respectively. CD4 T cell reconstitution was comparable among all chimeras but was lacking in the control WT→CIITA−/− chimeric mice (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070321/DC1). Total splenic CD4 T cells from the BM chimeras were skewed under Th1- or Th2-inducing conditions, and their cytokine-producing capacities were assessed by ICS. Th1 cells derived from CIITATg→B6 and CIITATg→CIITA−/− BM chimeric mice produced abundant IL-4 in addition to IFN-γ, with a consistently higher percentage of IL-4–producing Th1 cells from CIITATg→CIITA−/− mice (Fig. 3 A). In addition, the Th2 cell cultures from CIITATg→B6 and CIITATg→CIITA−/− chimeras that had T-CD4 T cells also produced greater numbers of IFN-γ–expressing cells than WT→B6 mice, whereas IFN-γ production by Th2 cells was variable among chimeras (Fig. 3 A). This variability in cells producing IFN-γ alone or IFN-γ together with IL-4 in WT→B6 mice seems to correlate with cell activation caused by the lymphopenic environment of the hosts (19) because these two populations are not present in WT mice (Fig. 2 B).
Figure 3.
The CD4 T cell selection pathway is responsible for IL-4–producing Th1 cells. (A) WT→B6, CIITATg→B6, and CIITATg→CIITA−/− chimeric mice were generated as described in Materials and methods. 11 wk after reconstitution, splenic CD4 T cells were differentiated under Th1- or Th2-inducing conditions for 6 d. Differentiated cells were then restimulated with PMA and ionomycin and analyzed for IFN-γ and IL-4 production. (B) CIITA transgene expression in the absence of MHC class II cannot generate IL-4–producing Th1 cells. BM from WT, CIITATg, or CIITATg/Aβ−/− mice were transplanted into lethally irradiated B6 mice. Differentiated Th1 or Th2 cells from the mice reconstituted for 3 mo were assayed for cytokine production by ICS. The percentage of positive cells in each quadrant is shown.
It was possible that the Th2 cytokine production by Th1 cells and enhanced IFN-γ production by Th2 cells from CIITATg→B6 and CIITATg→CIITA−/− BM chimeric mice was caused by CIITA expression rather than thymic selection differences. To determine whether CIITA expression itself regulates cytokine gene expression or whether thymic selection is indeed responsible for the phenotype, we tested BM from CIITATg/Aβ−/− mice. Cells from CIITATg/Aβ−/− mice express the CIITA transgene but not MHC class II because of a deficiency in the MHC class II structural gene Aβ. Thus, CIITATg/Aβ−/− BM-originated thymocytes in the chimera cannot mediate T-CD4 T cell selection because they do not express MHC class II molecules (18, 19). As a consequence, all CD4 T cells developed in CIITATg/Aβ−/−→B6 chimeras are selected on cTECs. If thymic selection determines cytokine expression potential, the resulting CIITATg/Aβ−/− Th1 and Th2 cells from the chimeric mice should not express IL-4 and IFN-γ, respectively. As shown in Fig. 3 B, the cytokine pattern of Th1 cells from CIITATg/Aβ−/−→B6 chimeras was indistinguishable from that of WT→B6 chimeras. Therefore, the thymocyte-mediated T cell selection pathway, not CIITA expression per se, is responsible for the generation of IL-4–producing Th1 cells and IFN-γ–producing Th2 cells.
Antigen presentation potential of TECs versus thymocytes dictates selection and the phenotype of the resulting CD4 T cells
We previously reported that MHC class II I-E expressed as a transgene under the control of the MHC class I promoter (I-ETg; Fig. S1 A) can also mediate CD4 T cell selection in the absence of endogenous CIITA, albeit at a low efficiency (19). Interestingly, CD4 T cells in these mice (I-ETg/CIITA−/−) also showed a similar phenotype to that of CD4 T cells in CIITATg mice (Table I) (31). Surprisingly, however, the same I-E transgene on the Aβ−/− background (I-ETg/Aβ−/−) did not generate CD4 T cells that can produce IL-4 under Th1 cell differentiation conditions (Table I) (32). In both types of I-ETg mice, CD4 T cells are expected to be selected on thymocytes and, therefore, their phenotype should be similar. We wondered whether thymocytes and TECs in the two models have a differential potential to support CD4 T cell selection, which results in the different phenotype of CD4 T cells. Unlike the MHC class II I-E transgene, CIITA also induces the expression of Ii and H-2M in cells, which allow them to become efficient APCs (33, 34). In fact, CIITATg thymocytes express high levels of both Ii and H-2M mRNA (unpublished data). Conversely, CIITA−/− cells express decreased Ii and H-2M (35). Therefore, introducing the I-E transgene to CIITA−/− mice cannot restore the APC function of TECs at full capacity, whereas the same transgene expression in Aβ−/− TECs can. Likewise, thymocytes expressing CIITA would be more efficient APCs than those expressing I-E only. Together with the different phenotype of I-ETg mice on the Aβ−/− or CIITA−/− background, it appears that the potency of antigen presentation of thymocytes and TECs dictates the direction of CD4 T cell selection toward the T- or E-CD4 T cell lineage.
Table I.
Relationship of the potency of antigen presentation in BM chimeras and the Th cell phenotype
| Mice | MHC II expression
|
Ii and H-2M
|
||||
|---|---|---|---|---|---|---|
| TECs | Thymocytes | TECs | Thymocytes | Predicted potency of antigen presentation |
Th2 cytokine production by Th1 cells |
|
| B6 | + | − | + | − | TEC | N |
| CIITATg | + | + | + | + | TEC = thy | Y |
| CIITATg/CIITA−/− | − | + | − | + | thy | Y |
| I-ETg | + | + | + | − | TEC > thy | N |
| I-ETg/Aβ−/− | + | + | + | − | TEC > thy | N |
| I-ETg/CIITA−/− | + | + | − | − | TEC = thy | Y |
| CIITATg→B6 | + | + | + | + | TEC = thy | Y |
| CIITATg→Aβ−/− | − | + | + | + | thy | Y |
| CIITATg→CIITA−/− | − | + | − | + | thy | Y |
| CIITATg/Aβ−/−→B6 | + | − | + | + | TEC | N |
| I-ETg→B6 | + | + | + | − | TEC > thy | N |
| I-ETg→Aβ−/− | − | + | + | − | thy | Y |
| I-ETg/CIITA−/−→B6 | + | + | + | − | TEC > thy | N |
| I-ETg/CIITA−/−→Aβ−/− | − | + | + | − | thy | Y |
Mice indicated with → are BM chimeras. Cells expressing MHC class II, Ii, and H-2M are considered hypothetically more potent than the cells expressing MHC class II in the absence or in low levels of Ii and H-2M. Expression denoted with − includes cells that express none or a low level of the indicated molecules. = or > show an equivalent or a stronger potency of antigen presentation function, respectively. Th2 cytokine production was experimentally determined using ICS. N, no; Y, yes.
To address this hypothesis, we constructed and tested several additional BM chimeras. The expression pattern of MHC class II, Ii, and H-2M in TECs and thymocytes in mice used in those experiments is shown in the Table S1 (available at http://www.jem.org/cgi/content/full/jem.20070321/DC1). As summarized in Table I, the results showed that if the predicted potency of thymocytes being APCs is greater than TECs (CIITATg/CIITA−/−, CIITATg→Aβ−/−, CIITATg→CIITA−/−, I-ETg→Aβ−/−, and I-ETg/CIITA−/−→Aβ−/−) or equivalent to that of TECs (CIITATg, I-ETg/CIITA−/−, and CIITATg→B6), CD4 T cells seem to be selected on thymocytes as efficiently as TECs, and the resulting CD4 T cells can express Th2 cytokines under Th1-inducing conditions. In contrast, CD4 T cells did not show the same phenotype when they were developed in mice in which TECs have a greater potential to present antigens than thymocytes (B6, I-ETg, I-ETg/Aβ−/−, CIITATg/Aβ−/−→B6, I-ETg→B6, and I-ETg/CIITA−/−→B6). Therefore, these data clearly demonstrate a strong correlation between thymocyte-mediated selection and the Th cell function of the resulting CD4 T cells.
Thymocyte-mediated selection pathway can override a requirement of Stat6 in Th2 cytokine expression
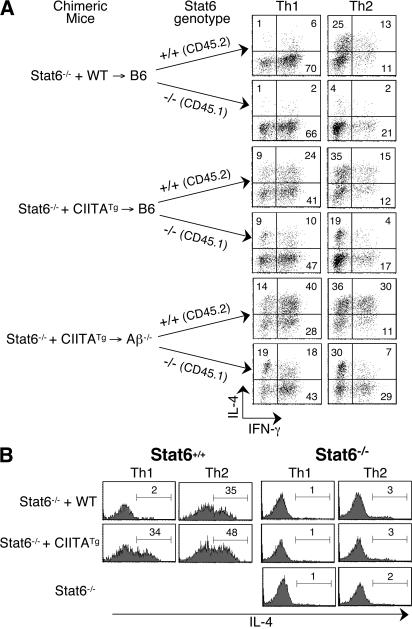
NKT cells that are selected on thymocytes produce IL-4 in a Stat6-independent manner (36, 37). Therefore, we asked whether IL-4 production by T-CD4 T cells requires Stat6. We have previously observed that CD4 T cells from CIITATg mice bred to Stat6−/− mice produced IL-4 (29). However, those data did not distinguish whether Stat6 independency is caused by thymic selection. Thus, we tested Stat6−/− CD4 T cells for their ability to produce IL-4 after being selected on thymocytes. For this experiment, we mixed two different sources of BM, as MHC class II–positive thymocytes can mediate the development of MHC class II–negative thymocytes in mixed BM chimeras (unpublished data) (18). Stat6−/− BM, together with WT or CIITATg (Stat6+/+) BM, were cotransferred into B6 or Aβ−/− hosts. To identify different populations of CD4 T cells, we used the congenic marker CD45. Splenic CD4 T cells from reconstituted animals were skewed under Th1- and Th2-polarizing conditions, and Th1 cells derived from Stat6+/+ BM (CD45.2+) and Stat6−/− BM (CD45.1+) were analyzed for cytokine production by ICS. As expected, Stat6+/+ cells from Stat6−/− + WT→B6 chimeric mice expressed IL-4 when they were differentiated to Th2 but not Th1 cells (Fig. 4 A, top group). When Stat6−/− cells were examined, both Stat6−/− Th1 and Th2 cells from Stat6−/− + WT→B6 BM chimeras produced a negligible amount of IL-4. In contrast, if Stat6−/− were developed in the presence of CIITATg BM-derived thymocytes, Stat6−/− Th1 and Th2 CD4 T cells produced IL-4 (Fig. 4 A, middle and bottom groups). Moreover, Stat6−/− CD4 T cells that were exclusively selected on thymocytes (Stat6−/− + CIITATg→Aβ−/−) had a greater potential to express IL-4. Stat6+/+ cells made IL-4 after differentiation into either the Th1 or Th2 cell lineage when they were from both Stat6−/− + CIITATg→B6 and Stat6−/− + CIITATg→Aβ−/− hosts. The Th2 cell cultures generated more IL-4–expressing cells than the Th1 cells (Fig. 4 A). We also examined cytokine production by freshly isolated Stat6−/− thymic and splenic CD4 T cells from mixed BM chimeras. Again, whenever the thymocyte-mediated selection pathway was available, Stat6−/− cells acquired the potential to express IL-4 (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20070321/DC1).
Figure 4.
Stat6-independent production of IL-4 by T-CD4 T cells. (A) Mixed BM chimeras. BM from Stat6−/− mice were mixed with those from WT or CIITATg mice and cotransferred into B6 or Aβ−/− recipients. Splenic CD4 T cells from mixed BM chimeras were differentiated into Th1 or Th2 cells. Before fixation and permeabilization, Th cells were stained with an anti-CD45.2 mAb to distinguish T cells derived from Stat6−/− BM (CD45.1+) as opposed to those from Stat6+/+ BM (CD45.2+). Cytokine production profiles were subsequently assayed by ICS. Mice were analyzed 10 wk after BM transplantation. The percentage of positive cells in each quadrant is shown. (B) Co-culturing Stat6−/− CD4 T cells with CIITATg CD4 T cells during in vitro Th cell differentiation is not sufficient to induce IL-4 expression. Histograms show IL-4 production profiles of Stat6+/+ WT or CIITATg cells (CD45.1+), or Stat6−/− cells (CD45.2+) in the co-cultures. The percentage of positive cells in each gate is shown.
Because Stat6−/− CD4 T cells were differentiated together with Stat6+/+ CD4 T cells, it was possible that the IL-4 produced by CIITATg CD4 T cells could have skewed Stat6−/− CD4 T cells to produce IL-4. To test this possibility, purified splenic CD4 T cells from Stat6−/− mice were mixed in vitro with WT or CIITATg CD4 T cells and differentiated under Th1- and Th2-inducing conditions. Stat6+/+ cells showed the expected cytokine profile from both WT and CIITATg Th1 and Th2 cells (Fig. 4 B). However, Stat6−/− CD4 T cells that were codifferentiated with CIITATg CD4 T cells did not acquire the ability to express IL-4. (Fig. 4 B). Collectively, our data strongly suggest that T-CD4 T cells produce Th2 cytokines in a Stat6-independent manner.
NKT cells do not influence the phenotype of T-CD4 T cells
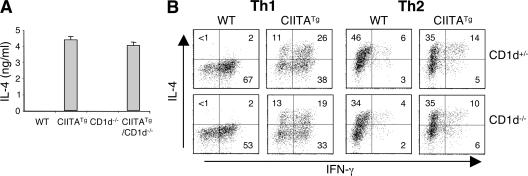
The rapid production of IFN-γ and IL-4, the Th0-like phenotype of T-CD4 cells, and the Stat6-independent IL-4 production by T-CD4 cells seem to be similar to that of NKT cells (36, 37). NKT cells play an important role in regulating both innate and adaptive immune responses through their prompt production of large amounts of cytokines, including IL-4 upon both in vivo and in vitro TCR stimulation. Therefore, we investigated whether NKT cells contribute to IL-4 expression in T-CD4 T cells using CIITATg mice lacking CD1d (CIITATg/CD1d−/−), which is necessary for NKT cell development (38). Primarily, when in vivo cytokine production was analyzed, a lack of CD1d and thus a deficiency in NKT cells did not alter the ability of CIITATg mice to produce IL-4 in response to short in vivo stimulation (Fig. 5 A). In addition, CD1d deficiency did not compromise the IL-4 production by CIITATg Th1 cells, thymocytes, and peripheral CD4 T cells upon in vitro stimulation (Fig. 5 B and not depicted). Therefore, CD1d-restricted NKT cells do not contribute to the Th2 cytokine–producing potential of T-CD4 T cells.
Figure 5.
NKT cells are not responsible for the IL-4–producing potential of T-CD4 Th1 cells. (A) Age-matched WT, CIITATg, CD1d−/−, and CIITATg/CD1d−/−mice were injected with 10 μg anti-CD3 antibody and killed 2 h later. The circulating IL-4 level in the serum was determined by ELISA. The error bars represent the mean ± SD of IL-4 measurements in the indicated mice. (B) CD4 T cells from CIITATg mice that were sufficient or deficient in CD1d expression were examined for their cytokine production profile by ICS after Th1 or Th2 cell differentiation. WT or CIITATg littermates on the CD1d+/− background were used as controls. Data are representative of at least two independent experiments. The percentage of positive cells in each quadrant is shown.
Preformed IL-4 mRNA in T-CD4 cells
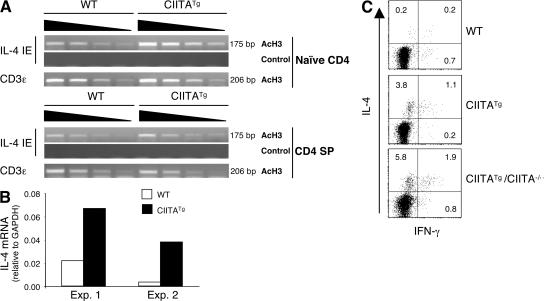
IL-4 expression in freshly isolated CIITATg CD4 T cells (Fig. S5) (29) suggested that the chromatin structure at the IL-4 locus might already be differentially modified. To test this, we performed a chromatin immunoprecipitation (ChIP) assay to analyze the acetylation status of histone H3 at the IL-4 intronic enhancer (IE) that has been shown to be hyperacetylated in differentiated Th2 cells (39–41). Naive CD4 T cells were purified from total peripheral CD4 cells obtained from WT and CIITATg mice by FACS sorting and subjected to the ChIP assay. In naive CD4 T cells, histone H3 at this regulatory region was indeed hyperacetylated compared with the WT cells (Fig. 6 A, top). The modified IL-4 locus in naive cells raised the possibility that a similar change might have occurred in the thymus. Indeed, purified CD4 SP thymocytes from CIITATg mice showed a small but consistent increase in H3 acetylation (Fig. 6 A, bottom). Moreover, this small change correlated with IL-4 gene transcription even in the absence of stimulation, as the amount of IL-4 transcripts was greater in freshly isolated CIITATg CD4 SP cells than in control cells (Fig. 6 B). The level of IFN-γ mRNA in those CIITATg thymocytes was variable and did not show a consistent increase (unpublished data). Consistent with the RNA data, NK1.1− CD4 SP cells produced a small but reproducibly higher percentage of IL-4–expressing cells in CIITATg and CIITATg/CIITA−/− but not WT mice (Fig. 6 C). The IL-4 producers were all CD44hi, which is similar to IL-4–producing thymic NKT cells (unpublished data). We also examined Stat6−/− thymocytes prepared from mixed BM chimeric mice and found that Stat6−/− thymocytes produced IL-4 when they were selected on thymocytes (Fig. S5 A). Collectively, our results indicate that the IL-4 locus in CD4 T cells that are selected on thymocytes is programmed for increased expression before Th cell differentiation.
Figure 6.
T-CD4 T cells acquire the potential to express IL-4 in the thymus. (A) Histone H3 at the IL-4 locus is hyperacetylated in CIITATg CD4 T cells and thymocytes. Naive (CD44loCD45RBhi) CD4 T cells and CD4 SP thymocytes from WT and CIITATg mice were sorted and used for the ChIP assay with an antiacetylated histone H3 antibody. PCR primers specific for the IL-4 enhancer were used to amplify the precipitated DNA. No antibody was used as the negative control for the immunoprecipitation, and primers specific for CD3ε were used as an internal loading control. Note that the CD3ε mRNA level was equivalent between WT and CIITATg T cells (not depicted). (B) CIITATg CD4 SP thymocytes are poised to express the IL-4 gene. RNA was extracted from CD4 SP thymocytes from WT and CIITATg mice immediately after FACS sorting. IL-4 mRNA was quantified by quantitative RT-PCR, and the results were expressed as ratios relative to the housekeeping gene GAPDH. (C) CD4 SP T cells are capable of producing IL-4. Sorted NK1.1− CD4 SP cells from WT, CIITATg, or CIITATg/CIITA−/− thymocytes were stimulated with PMA plus ionomycin for 5 h. Cytokine production was analyzed by ICS. The percentage of positive cells in each quadrant is shown.
T-CD4 T cells can dampen allergen-induced airway inflammation
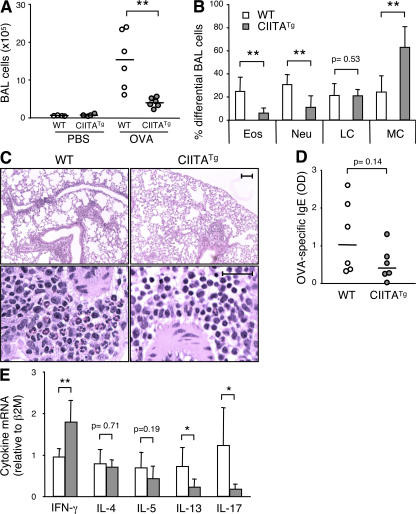
To investigate what role T-CD4 T cells play during an immune response in vivo, we used an antigen-induced airway inflammation model of asthma. CIITATg and WT mice were sensitized with chicken OVA and subsequently challenged with aerosolized OVA to induce inflammation in the respiratory tract. Mice were then killed to measure several parameters that are indicative of airway inflammation. We first examined the number of total cells in the bronchoalveolar lavage fluid (BALF). As shown in Fig. 7 A, the total cell numbers in BALF from OVA-challenged mice were decreased in CIITATg mice. Infiltration of eosinophils in the lung tissue is one of the cardinal features of asthma and serves as a simple readout for airway inflammation. In agreement with the total BALF cell numbers, CIITATg mice had significantly fewer BALF eosinophils than WT mice (Fig. 7 B). Neutrophilic infiltration in BALF was also reduced in CIITATg mice (Fig. 7 B). The percentage of monocytes was significantly higher in CIITATg than in WT mice, whereas the level of lymphocytes was comparable between WT and CIITATg mice (Fig. 7 B). Hematoxylin and eosin (H&E) staining of lung tissue sections also supported the difference in eosinophil infiltration between the two groups of mice (Fig. 7 C). In addition, OVA-specific IgE levels in serum were decreased in CIITATg mice (Fig. 7 D). We further examined the expression of several cytokines involved in airway inflammation. The lung tissue from OVA-sensitized and -challenged CIITATg mice had significantly more IFN-γ but less IL-13 and IL-17 transcripts compared with the WT control mice. The IL-4 and IL-5 mRNA levels were comparable between the two groups (Fig. 7 E). Collectively, T-CD4 T cells seem to play a protective role during airway inflammation, which was associated with a distinct Th effector cytokine production profile at the site of inflammation.
Figure 7.
Allergen-induced airway inflammation is attenuated in CIITATg mice. Total BALF cells (A) and percentages of differential cell counts (B) in the BALF of WT and CIITATg mice 48 h after the last OVA aerosol treatment were determined. Each symbol in the graphs represents one mouse. Eos, eosinophils; Neu, neutrophils; LM, lymphocytes; MC, monocytes. (C) Reduced perivascular eosinophilic infiltration in CIITATg lung sections 2 d after the last exposure to OVA. Eosinophils are the red-staining cells in the H&E lung sections. Bars: (top) 100 μm; (bottom) 25 μm. (D) OVA-specific serum IgE levels. (E) Real-time RT-PCR of cytokine mRNA levels in the lung tissue of the OVA-challenged mice. The error bars represent the mean ± SD of six mice in each group. Horizontal lines in A and D represent median values. The Student's two-tailed t test was used to calculate statistical significance. Data are pooled from two independent experiments. P < 0.05 was considered statistically significant. *, P < 0.05; **, P < 0.01.
DISCUSSION
In this study, we have shown that the thymic selection process plays an important role in the cytokine production potential of CD4 T cells. Hence, unlike E-CD4 T cells, T-CD4 T cells secrete both Th1 and Th2 cytokines shortly after stimulation. Remarkably, IL-4–producing cells can develop in the absence of Stat6 when CD4 T cells are selected on thymocytes. In addition, T-CD4 T cells can produce IL-5 and IL-13 in the absence of IL-4 (29). These observations are consistent with our data showing that the IL-4 locus, possibly including the IL-5 and IL-13 loci, is already remodeled in T-CD4 T cells such that the IL-5 and IL-13 genes are readily transcribed without IL-4.
Currently, it is not known how the thymic selection process regulates Th cell fate. Many factors are known to influence Th cell differentiation of conventional CD4 T cells, and the IL-4–Stat6 signaling pathway is critical for the differentiation of Th2 effector cells (14–17, 42–44). Stat6 activates GATA3, the master regulator of Th2 cells, which subsequently mediates chromatin remodeling accompanied by an increased level of histone hyperacetylation at the IL-4 locus. Although we could detect enhanced histone H3 and H4 acetylation at the IL-4 IE in both CD4 SP thymocytes and naive CD4 T cells from CIITATg mice, we found little difference in GATA3 expression in CIITATg cells as compared with WT controls (unpublished data). Consistent with this finding, we showed that Th2 cytokine production by T-CD4 T cells does not totally depend on the Stat6 signaling pathway. Interestingly, this Stat6-independent production of IL-4 is shared by NKT cells (36, 37). NKT cells coexpress surface markers characteristic of both conventional T cells and NK cells and are positively selected by nonclassic MHC class I molecule CD1d on cortical double-positive thymocytes (45–47). Functionally, NKT cells resemble innate effector cells in that they promptly produce large amounts of cytokines such as IFN-γ and IL-4 cytokines upon TCR stimulation. It was shown that during thymic development, the IFN-γ and IL-4 loci in NKT cells are modified by histone acetylation and both genes are constitutively transcribed, which correlates well with the capacity of NKT cells to rapidly produce cytokines (37). However, T-CD4 T cells are distinct from NKT cells (19), and their phenotype is not influenced by CD1d-restricted NKT cells.
T cells selected on thymocytes such as NKT cells and nonclassical MHC class Ib–restricted CD8 cells appear to have TCR with higher avidity (48–50). Similarly, it is possible that thymocytes with higher TCR avidity could have been preferentially selected on MHC class II–positive thymocytes. The TCR signaling potency is known to be associated with Th cell differentiation (51–55). Therefore, the difference in TCR signaling potency could contribute to Th cytokine production by T-CD4 T cells. Previously, we demonstrated that the antigen repertoire of thymocytes does not totally overlap with that of TECs (19). Our current data also showed that the antigen presentation potential of thymic APCs seems to regulate the selection pathway and Th cell fate. Therefore, it appears that developing thymocytes receive different signal strength when they are selected on thymocytes, generating a distinct cellular phenotype of cells. This is consistent with the similar cytokine production potential observed in both T-CD4 T cells and NKT cells.
Despite the fact that T-CD4 cells can readily express Th2 cytokines in vivo, mice that can generate T-CD4 T cells were less susceptible to airway inflammation induced by a Th2-biased allergen. Although it is not yet clear how T-CD4 T cells exert this suppressive effect, several possibilities can be envisioned. First, T-CD4 T cells can produce IFN-γ at a high level upon stimulation. Because IFN-γ has been shown to suppress the development of mouse airway inflammation (56, 57), it is possible that the IFN-γ produced by T-CD4 T cells can dampen Th2-inducing conditions in vivo, which in turn may attenuate the function of Th2 cells during disease development. Second, there was a marked reduction of IL-17 transcripts in the lung tissue of the OVA-challenged CIITATg mice. IL-17 has been shown to play an important role in many immune diseases, including airway inflammation (3, 58, 59). Because both IL-4 and IFN-γ suppress the generation of Th17 (2, 3), T-CD4 T cells with a dual cytokine production potential might have altered Th17 cell differentiation, contributing to protection. Indeed, CIITATg CD4 T cells make substantially less IL-17 than WT CD4 T cells in vitro (unpublished data). Reduced IL-17 production in CIITATg mice could also be responsible for protecting them from experimental autoimmune encephalomyelitis (60). Third, T-CD4 T cells may have a limited capability to expand during the challenge period, and mount an inefficient memory response. In this regard, nonclassical MHC class Ib–restricted CD8 cells that can be selected on TECs or hematopoietic cells failed to undergo secondary expansion upon Listeria infection, even though they can mount a rapid primary immune response (50, 61–63). Lastly, it is possible that OVA-specific effector T-CD4 T cells could not be generated efficiently or their infiltration into the lung was defective. An identification of markers specific for T-CD4 T cells would help to address this issue in the future. Regardless of mechanism, T-CD4 T cells appear to play a critical and diverse role during an immune response.
A substantial portion of human fetal and neonatal thymocytes express MHC class II molecules, which have been shown to be involved in the positive selection of CD4 T cells through interactions between thymocytes (64). Therefore, it is reasonable to propose that healthy individuals have two CD4 T cell populations selected on TECs as well as thymocytes. At the moment, nothing is known about the regulation of these two pathways. We do not know whether the two populations of CD4 T cells are selected and maintained equally or whether one population is preferentially selected over the other. Nonetheless, it is conceivable that the differential selection efficiency in an individual could have profound consequences in immunity. Whereas Th cell differentiation is relatively easy to observe in mice, the paradigm has not been as clear in the human system (65–70). In addition, differentiated human Th cells can produce both cytokines when they are cultured in the opposing polarizing condition (71), whereas committed mouse Th cells cannot be reversed (72). The phenotype of human Th cells suggests a distinct genetic programming in their cytokine gene expression, which appears to be similar to T-CD4 T cells in CIITATg mice. Perhaps the presence of two selection pathways in humans is responsible for generating CD4 T cells that produce both IL-4 and IFN-γ. Further investigations are warranted to elucidate the role of T-CD4 T cells in the modulation of immune diseases in humans.
MATERIALS AND METHODS
Mice.
Mice carrying the human type III CIITA transgene (CIITATg), CIITA-deficient mice (CIITA−/−), CIITATg mice on the CIITA-deficient background (CIITATg/CIITA−/−) and on the MHC class II–deficient Aβ−/− background (CIITATg/Aβ−/−), MHC class II I-E transgenic mice (I-ETg), I-ETg mice on the CIITA-deficient background (I-ETg/CIITA−/−) and on the MHC class II–deficient Aβ−/− background (I-ETg/Aβ−/−), and Stat6−/− mice were previously described (19, 29, 32, 34, 44). CD1d-deficient mice on the C57BL/6 background (38) were bred with CIITATg mice to generate CIITATg/CD1d−/−mice, and WT and CIITATg/CD1d+/− littermates were used as CD1d-positive controls. Stat6−/− mice were bred onto the H-2b background and carried the CD45.1 congenic marker. C57BL/6 (B6) mice and the MHC class II–deficient Aβ−/− mice were purchased from the Jackson Laboratory and Taconic, respectively, and bred in the animal facility at the Indiana University School of Medicine (IUSM). All mice were housed under specific pathogen-free conditions and used at 6–12 wk of age. All animal experiments were performed under protocols approved by the IUSM Animal Care and Use Committee.
In vivo anti-CD3 stimulation.
PBS or 10 μg anti-CD3 antibody in PBS was injected into WT or CIITATg mice through the tail vein. 2 h later, mice were killed and blood was collected by cardiac puncture. Serum cytokines were determined by ELISA.
BM chimeras.
Recipient B6, Aβ−/−, or CIITA−/− mice were lethally irradiated (950 rad) and rested for 24 h before receiving BM cells. Total BM cells were prepared from the femurs and tibias of donor mice (2–3 mo of age) and depleted of mature T cells, B cells, and MHC class II–positive lymphocytes by using a cocktail of antibodies containing anti-CD4 (RL172) and anti-CD8 (TIB105, TIB210), anti-CD19 (1D3), and anti–MHC class II (M5/114), followed by complement-mediated lysis. These cells were subsequently referred to as T-depleted BM cells. Each recipient mouse received 2.5 × 106 T-depleted BM cells in 500 μl of 1× PBS via tail vein injection. Reconstituted mice were analyzed 2–3 mo later.
Flow cytometry.
All antibodies for flow cytometry were purchased from BD Biosciences, and cells were preincubated with the anti-FcγR mAb 2.4G2 to block nonspecific antibody binding. The following FITC-, PE-, PerCP-, cychrome-, allophycocyanin-, or biotin-conjugated antibodies were used: CD4 (L3T4), CD8 (53-6.7), CD45RB (16A), CD44 (IM7), NK1.1 (PK136), CD1d (1B1), CD45.1 (A20), CD45.2 (104), anti–IL-4 (11B11), and anti–IFN-γ (XMG1.2). Allophycocyanin-conjugated streptavidin was used to visualize staining by biotinylated primary antibodies. Events were acquired on a flow cytometer (FACSCalibur; Beckman Dickinson), and the data were analyzed using CellQuest software (BD Biosciences).
T cell preparation and stimulation.
To purify CD4 and CD8 SP T cells, total thymocytes were depleted of double-positive cells by complement-mediated lysis of HSA+ cells. The remaining cells were subsequently sorted electronically for NK1.1−CD4+CD8− and NK1.1−CD4−CD8+ cells. Total peripheral CD4 cells were enriched from single-cell suspensions from spleen and lymph nodes (auxiliary, brachial, inguinal, and mesenteric) with anti–mouse CD4 microbeads (Miltenyi Biotec). To obtain naive CD4 T cells, enriched CD4 T cells were stained with anti-CD4, CD45RB, and CD44 and electronically sorted for CD4+CD45RBhiCD44lo cells. In some experiments, CD4 T cells were enriched from splenocytes. To induce Th cell differentiation under neutral conditions (ThN), total peripheral, splenic CD4 T cells or 106 naive T cells/ml were stimulated with 5 μg/ml of plate-bound anti-CD3ε (145-2C11), 1 μg/ml anti-CD28 (37.51), and 50 U IL-2 (Roche) for 5–7 d. For Th1 cell differentiation, and additional 3.5 ng/ml IL-12 and 10 μg/ml anti–IL-4 (11B11) were added. Th2 cell cultures were supplemented with 10 ng/ml IL-4 and 10 μg/ml anti–IFN-γ (R4-6A2).
Cytokine assays.
For the ELISA assays, differentiated cells were restimulated overnight with 5 μg/ml of plate-bound anti-CD3 antibody at a cell density of 1 × 106/ml. IL-4 and IFN-γ in the supernatants were quantified by paired cytokine-specific antibodies (BD Biosciences). Recombinant cytokines were used as standards. For ICS, cells were stimulated with 50 ng/ml phorbol myristyl acetate and 1.5 μM ionomycin (Calbiochem) for 5 h. Monensin (Sigma-Aldrich) at 3 μM was added during the last 3 h of stimulation. Activated cells were stained with anti-CD4 and anti-NK1.1 in some cases. Cells were fixed in 2–4% paraformaldehyde and permeabilized with 0.2% saponin (Sigma-Aldrich), followed by staining with anti-IL4 (11B11) and anti–IFN-γ (XMG1.2) for flow cytometry.
RT-PCR.
Total RNA was extracted from sorted CD4 SP thymocytes lysed in TRIzol reagent (Invitrogen), according to the manufacturer's recommendations, and reverse transcribed using the SuperScript First-Strand cDNA Synthesis System (Invitrogen). Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). All PCR reactions were done in triplicate, and the data were analyzed by the comparative threshold cycle (ΔCT) method and normalized to GAPDH. The primer pairs used for GAPDH were 5′-CCAGGTTGTCTCCTGCGACT-3′ and 5′-ATACCAGGAAATGAGCTTGACAAAGT-3′, and for IL-4 were 5′-ACAGGAGAAGGGACGCCAT-3′ and 5′-GAAGCCCTACAGACGAGCTCA-3′ (73). Message levels of IFN-γ, IL-4, IL-5, IL-13, and IL-17A in the lung tissue were analyzed by TaqMan PCR (all reagents were obtained from Applied Biosystems). Cycle number of duplicate samples was normalized to expression of β2-microglobulin. Results are relative to one of the WT mice.
ChIP analysis.
ChIP analysis was performed according to the ChIP assay protocol (Upstate Biotechnology). In brief, 2–3 × 106 sorted CD4 or CD8 SP thymocytes or sorted naive CD4 cells from WT and CIITATg mice were fixed in 1% formaldehyde for 10 min at room temperature, washed, lysed, and sonicated with three pulses to generate chromatin fragments of ∼500-bp in length. Antiacetylated histone H3 antibody (Upstate Biotechnology) was added (3 μl per immunoprecipitation) to the diluted lysates and incubated overnight. No antibody group was used as a negative control. Protein A–sepharose CL-4B beads (GE Healthcare) were added for 1 h. After washes, the immunocomplexes were eluted, the cross-links were reversed, and DNA was purified by phenol/chloroform extraction and resuspended in 50 μl TE buffer. Semiquantitative PCR was done with twofold serial dilutions of ChIP DNA samples. The primers used for IL-4 IE were 5′-GGGTGTGAATAAGCCATATTG-3′ and 5′-CCCAGCGTTTACATGAGC-3′ (40), and for CD3ε were 5′-CATTTCCAAGTGACGTGG-3′ and 5′-AACACACTGGCTGCATGC-3′ (39).
OVA-induced airway inflammation.
On days 0 and 7, mice were sensitized i.p. with 20 μg OVA (grade V; Sigma-Aldrich) adsorbed onto 0.5 mg of aluminum hydroxide gel (Sigma-Aldrich) in 0.5 ml PBS. Beginning on day 14, mice were challenged with aerosolized OVA from a 1% OVA/PBS solution for 20 min using a jet nebulizer for 7 consecutive days. 2 d after the last aerosol challenge, mice were anesthetized with sodium pentobarbital. BALF was collected from airways by three washes with 1× PBS. After BALF collection, blood was drawn by a cardiac puncture, and a portion of the lung was fixed in 10% buffered formalin for histological examination. The remaining lung tissue was stored in RNAlater solution (Ambion) and homogenized in TRIzol for RNA preparation. After cytospin of the BALF, slides were stained with the Diff-Quick stain set (Baxter), and differential cell numbers were determined by counting 200 cells per slide. OVA-specific IgE levels in serum were measured by ELISA, and the data were expressed as OD readings. Fixed lung tissues were embedded in paraffin, cut into 5-μm-thick sections, and stained with H&E for histological analysis of airway inflammation.
Online supplemental material.
Table S1 shows MHC class II, Ii, and H-2M expression in mice used in this study. Fig. S1 shows MHC class II expression on thymocytes and CD44 and CD45RB expression on splenic CD4 T cells from WT, CIITATg, CIITATg/CIITA−/−, and I-ETg mice. Fig. S2 shows the gate used to sort naive CD4 T cells (CD45RBhiCD44lo). Fig. S3 shows IL-4 and IFN-γ production by Th1 and Th2 cells differentiated from total WT, CIITATg, and CIITATg/CIITA−/− CD4 T cells. Fig. S4 shows the generation of E- and T-CD4 T cells in BM chimeric mice. The FACS plots represent profiles of CD4 versus CD8 in the thymus and LN. Fig. S5 shows IL-4 production by TEC- and thymocyte-selected Stat6−/− thymocytes and splenic CD4 T cells from chimeras that were reconstituted with Stat6−/− and WT or CIITATg BM. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070321/DC1.
Supplemental Material
Acknowledgments
This work was supported in part by National Institutes of Health grant AI 45811.
The authors have no conflicting financial interests.
Abbreviations used: BALF, bronchoalveolar lavage fluid; ChIP, chromatin immunoprecipitation; CIITA, MHC class II transactivator; E-CD4, epithelial cell–selected CD4; cTEC, cortical TEC; H&E, hematoxylin and eosin; ICS, intracellular cytokine stating; IE, intronic enhancer; SP, single positive; T-CD4, thymocyte-selected CD4; TEC, thymic epithelial cell.
References
- 1.Mosmann, T.R., H. Cherwinski, M.W. Bond, M.A. Giedlin, and R.L. Coffman. 1986. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 175:5–14. [PubMed] [Google Scholar]
- 2.Harrington, L.E., R.D. Hatton, P.R. Mangan, H. Turner, T.L. Murphy, K.M. Murphy, and C.T. Weaver. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132. [DOI] [PubMed] [Google Scholar]
- 3.Park, H., Z. Li, X.O. Yang, S.H. Chang, R. Nurieva, Y.H. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian, and C. Dong. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver, C.T., L.E. Harrington, P.R. Mangan, M. Gavrieli, and K.M. Murphy. 2006. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 24:677–688. [DOI] [PubMed] [Google Scholar]
- 5.Langrish, C.L., Y. Chen, W.M. Blumenschein, J. Mattson, B. Basham, J.D. Sedgwick, T. McClanahan, R.A. Kastelein, and D.J. Cua. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cua, D.J., J. Sherlock, Y. Chen, C.A. Murphy, B. Joyce, B. Seymour, L. Lucian, W. To, S. Kwan, T. Churakova, et al. 2003. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 421:744–748. [DOI] [PubMed] [Google Scholar]
- 7.Yen, D., J. Cheung, H. Scheerens, F. Poulet, T. McClanahan, B. McKenzie, M.A. Kleinschek, A. Owyang, J. Mattson, W. Blumenschein, et al. 2006. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 116:1310–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwakura, Y., and H. Ishigame. 2006. The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 116:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington, L.E., P.R. Mangan, and C.T. Weaver. 2006. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr. Opin. Immunol. 18:349–356. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie, B.S., R.A. Kastelein, and D.J. Cua. 2006. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 27:17–23. [DOI] [PubMed] [Google Scholar]
- 11.Bettelli, E., Y. Carrier, W. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. [DOI] [PubMed] [Google Scholar]
- 12.Mangan, P.R., L.E. Harrington, D.B. O'Quinn, W.S. Helms, D.C. Bullard, C.O. Elson, R.D. Hatton, S.M. Wahl, T.R. Schoeb, and C.T. Weaver. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. [DOI] [PubMed] [Google Scholar]
- 13.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 14.Constant, S.L., and K. Bottomly. 1997. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu. Rev. Immunol. 15:297–322. [DOI] [PubMed] [Google Scholar]
- 15.Murphy, K.M., and S.L. Reiner. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944. [DOI] [PubMed] [Google Scholar]
- 16.Szabo, S.J., B.M. Sullivan, S.L. Peng, and L.H. Glimcher. 2003. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 21:713–758. [DOI] [PubMed] [Google Scholar]
- 17.Mowen, K.A., and L.H. Glimcher. 2004. Signaling pathways in Th2 development. Immunol. Rev. 202:203–222. [DOI] [PubMed] [Google Scholar]
- 18.Choi, E.Y., K.C. Jung, H.J. Park, D.H. Chung, J.S. Song, S.D. Yang, E. Simpson, and S.H. Park. 2005. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 23:387–396. [DOI] [PubMed] [Google Scholar]
- 19.Li, W., M.G. Kim, T.S. Gourley, B.P. McCarthy, D.B. Sant'angelo, and C.H. Chang. 2005. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 23:375–386. [DOI] [PubMed] [Google Scholar]
- 20.Saito, Y., Y. Kametani, K. Hozumi, N. Mochida, K. Ando, M. Ito, T. Nomura, Y. Tokuda, H. Makuuchi, T. Tajima, and S. Habu. 2002. The in vivo development of human T cells from CD34(+) cells in the murine thymic environment. Int. Immunol. 14:1113–1124. [DOI] [PubMed] [Google Scholar]
- 21.Yahata, T., K. Ando, Y. Nakamura, Y. Ueyama, K. Shimamura, N. Tamaoki, S. Kato, and T. Hotta. 2002. Functional human T lymphocyte development from cord blood CD34+ cells in nonobese diabetic/Shi-scid, IL-2 receptor gamma null mice. J. Immunol. 169:204–209. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu, H., R. Nishikomori, T. Heike, M. Ito, K. Kobayashi, K. Katamura, and T. Nakahata. 2003. Complete reconstitution of human lymphocytes from cord blood CD34+ cells using the NOD/SCID/gammacnull mice model. Blood. 102:873–880. [DOI] [PubMed] [Google Scholar]
- 23.Traggiai, E., L. Chicha, L. Mazzucchelli, L. Bronz, J.C. Piffaretti, A. Lanzavecchia, and M.G. Manz. 2004. Development of a human adaptive immune system in cord blood cell-transplanted mice. Science. 304:104–107. [DOI] [PubMed] [Google Scholar]
- 24.Klein, C., M. Cavazzana-Calvo, F. Le Deist, N. Jabado, M. Benkerrou, S. Blanche, B. Lisowska-Grospierre, C. Griscelli, and A. Fischer. 1995. Bone marrow transplantation in major histocompatibility complex class II deficiency: a single-center study of 19 patients. Blood. 85:580–587. [PubMed] [Google Scholar]
- 25.Godthelp, B.C., M.C. Van Eggermond, M.J. Van Tol, J.M. Vossen, and P.J. van den Elsen. 2000. T cell immune reconstitution after allogeneic bone marrow transplantation in bare lymphocyte syndrome. Hum. Immunol. 61:898–907. [DOI] [PubMed] [Google Scholar]
- 26.Markert, M.L., M. Sarzotti, D.A. Ozaki, G.D. Sempowski, M.E. Rhein, L.P. Hale, F. Le Deist, M.J. Alexieff, J. Li, E.R. Hauser, et al. 2003. Thymus transplantation in complete DiGeorge syndrome: immunologic and safety evaluations in 12 patients. Blood. 102:1121–1130. [DOI] [PubMed] [Google Scholar]
- 27.Markert, M.L., M.J. Alexieff, J. Li, M. Sarzotti, D.A. Ozaki, B.H. Devlin, D.A. Sedlak, G.D. Sempowski, L.P. Hale, H.E. Rice, et al. 2004. Postnatal thymus transplantation with immunosuppression as treatment for DiGeorge syndrome. Blood. 104:2574–2581. [DOI] [PubMed] [Google Scholar]
- 28.Rice, H.E., M.A. Skinner, S.M. Mahaffey, K.T. Oldham, R.J. Ing, L.P. Hale, and M.L. Markert. 2004. Thymic transplantation for complete DiGeorge syndrome: medical and surgical considerations. J. Pediatr. Surg. 39:1607–1615. [DOI] [PubMed] [Google Scholar]
- 29.Patel, D.R., W. Li, J.S. Park, M.H. Sofi, T.S. Gourley, G. Hangoc, M.H. Kaplan, and C.H. Chang. 2005. Constitutive expression of CIITA directs CD4 T cells to produce Th2 cytokines in the thymus. Cell. Immunol. 233:30–40. [DOI] [PubMed] [Google Scholar]
- 30.Reith, W., S. LeibundGut-Landmann, and J.M. Waldburger. 2005. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5:793–806. [DOI] [PubMed] [Google Scholar]
- 31.Patel, D.R., M.H. Kaplan, and C.H. Chang. 2004. Altered Th1 cell differentiation programming by CIITA deficiency. J. Immunol. 173:5501–5508. [DOI] [PubMed] [Google Scholar]
- 32.Gourley, T., S. Roys, N.W. Lukacs, S.L. Kunkel, R.A. Flavell, and C.H. Chang. 1999. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 10:377–386. [DOI] [PubMed] [Google Scholar]
- 33.Chin, K.C., C. Mao, C. Skinner, J.L. Riley, K.L. Wright, C.S. Moreno, G.R. Stark, J.M. Boss, and J.P. Ting. 1994. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1:687–697. [DOI] [PubMed] [Google Scholar]
- 34.Chang, C.H., and R.A. Flavell. 1995. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J. Exp. Med. 181:765–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang, C.H., S. Guerder, S.C. Hong, W. van Ewijk, and R.A. Flavell. 1996. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 4:167–178. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan, M.H., A.L. Wurster, S.T. Smiley, and M.J. Grusby. 1999. Stat6-dependent and -independent pathways for IL-4 production. J. Immunol. 163:6536–6540. [PubMed] [Google Scholar]
- 37.Stetson, D.B., M. Mohrs, R.L. Reinhardt, J.L. Baron, Z.E. Wang, L. Gapin, M. Kronenberg, and R.M. Locksley. 2003. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J. Exp. Med. 198:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mendiratta, S.K., W.D. Martin, S. Hong, A. Boesteanu, S. Joyce, and L. Van Kaer. 1997. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 6:469–477. [DOI] [PubMed] [Google Scholar]
- 39.Avni, O., D. Lee, F. Macian, S.J. Szabo, L.H. Glimcher, and A. Rao. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 3:643–651. [DOI] [PubMed] [Google Scholar]
- 40.Baguet, A., and M. Bix. 2004. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc. Natl. Acad. Sci. USA. 101:11410–11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fields, P.E., S.T. Kim, and R.A. Flavell. 2002. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol. 169:647–650. [DOI] [PubMed] [Google Scholar]
- 42.Shimoda, K., J. van Deursen, M.Y. Sangster, S.R. Sarawar, R.T. Carson, R.A. Tripp, C. Chu, F.W. Quelle, T. Nosaka, D.A. Vignali, et al. 1996. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 380:630–633. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, K., T. Tanaka, W. Shi, M. Matsumoto, M. Minami, S. Kashiwamura, K. Nakanishi, N. Yoshida, T. Kishimoto, and S. Akira. 1996. Essential role of Stat6 in IL-4 signalling. Nature. 380:627–630. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan, M.H., U. Schindler, S.T. Smiley, and M.J. Grusby. 1996. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 4:313–319. [DOI] [PubMed] [Google Scholar]
- 45.Kronenberg, M., and L. Gapin. 2002. The unconventional lifestyle of NKT cells. Nat. Rev. Immunol. 2:557–568. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi, M., M. Harada, S. Kojo, T. Nakayama, and H. Wakao. 2003. The regulatory role of Valpha14 NKT cells in innate and acquired immune response. Annu. Rev. Immunol. 21:483–513. [DOI] [PubMed] [Google Scholar]
- 47.Kronenberg, M. 2005. Toward an understanding of NKT cell biology: progress and paradoxes. Annu. Rev. Immunol. 23:877–900. [DOI] [PubMed] [Google Scholar]
- 48.Sidobre, S., O.V. Naidenko, B.C. Sim, N.R. Gascoigne, K.C. Garcia, and M. Kronenberg. 2002. The V alpha 14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J. Immunol. 169:1340–1348. [DOI] [PubMed] [Google Scholar]
- 49.Cantu, C., III, K. Benlagha, P.B. Savage, A. Bendelac, and L. Teyton. 2003. The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J. Immunol. 170:4673–4682. [DOI] [PubMed] [Google Scholar]
- 50.Urdahl, K.B., J.C. Sun, and M.J. Bevan. 2002. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat. Immunol. 3:772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosken, N.A., K. Shibuya, A.W. Heath, K.M. Murphy, and A. O'Garra. 1995. The effect of antigen dose on CD4+ T helper cell phenotype development in a T cell receptor–αβ–transgenic model. J. Exp. Med. 182:1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffer, C., J. Stein, S. Southwood, H. Ketelaar, A. Sette, and K. Bottomly. 1995. Altered peptide ligands can control CD4 T lymphocyte differentiation in vivo. J. Exp. Med. 181:1569–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Constant, S., C. Pfeiffer, A. Woodard, T. Pasqualini, and K. Bottomly. 1995. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J. Exp. Med. 182:1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tao, X., C. Grant, S. Constant, and K. Bottomly. 1997. Induction of IL-4-producing CD4+ T cells by antigenic peptides altered for TCR binding. J. Immunol. 158:4237–4244. [PubMed] [Google Scholar]
- 55.Rogers, P.R., C. Dubey, and S.L. Swain. 2000. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J. Immunol. 164:2338–2346. [DOI] [PubMed] [Google Scholar]
- 56.Cohn, L., R.J. Homer, N. Niu, and K. Bottomly. 1999. T helper 1 cells and interferon γ regulate allergic airway inflammation and mucus production. J. Exp. Med. 190:1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fulkerson, P.C., N. Zimmermann, E.B. Brandt, E.E. Muntel, M.P. Doepker, J.L. Kavanaugh, A. Mishra, D.P. Witte, H. Zhang, J.M. Farber, et al. 2004. Negative regulation of eosinophil recruitment to the lung by the chemokine monokine induced by IFN-gamma (Mig, CXCL9). Proc. Natl. Acad. Sci. USA. 101:1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakae, S., Y. Komiyama, A. Nambu, K. Sudo, M. Iwase, I. Homma, K. Sekikawa, M. Asano, and Y. Iwakura. 2002. Antigen-specific T cell sensitization is impaired in IL-17-deficient mice, causing suppression of allergic cellular and humoral responses. Immunity. 17:375–387. [DOI] [PubMed] [Google Scholar]
- 59.Linden, A. 2006. Interleukin-17 and airway remodelling. Pulm. Pharmacol. Ther. 19:47–50. [DOI] [PubMed] [Google Scholar]
- 60.Park, W.S., Y. Bae, D.H. Chung, Y.L. Choi, B.K. Kim, Y.C. Sung, E.Y. Choi, S.H. Park, and K.C. Jung. 2004. T cell expression of CIITA represses Th1 immunity. Int. Immunol. 16:1355–1364. [DOI] [PubMed] [Google Scholar]
- 61.Kerksiek, K.M., D.H. Busch, I.M. Pilip, S.E. Allen, and E.G.P. Am. 1999. H2-M3–restricted T cells in bacterial infection: rapid primary but diminished memory responses. J. Exp. Med. 190:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamilton, S.E., B.B. Porter, K.A. Messingham, V.P. Badovinac, and J.T. Harty. 2004. MHC class Ia-restricted memory T cells inhibit expansion of a nonprotective MHC class Ib (H2-M3)-restricted memory response. Nat. Immunol. 5:159–168. [DOI] [PubMed] [Google Scholar]
- 63.Bouwer, H.G., R.A. Barry, and D.J. Hinrichs. 2001. Lack of expansion of major histocompatibility complex class Ib-restricted effector cells following recovery from secondary infection with the intracellular pathogen Listeria monocytogenes. Infect. Immun. 69:2286–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi, E.Y., W.S. Park, K.C. Jung, D.H. Chung, Y.M. Bae, T.J. Kim, H.G. Song, S.H. Kim, D.I. Ham, J.H. Hahn, et al. 1997. Thymocytes positively select thymocytes in human system. Hum. Immunol. 54:15–20. [DOI] [PubMed] [Google Scholar]
- 65.Allen, J.E., and R.M. Maizels. 1997. Th1-Th2: reliable paradigm or dangerous dogma? Immunol. Today. 18:387–392. [DOI] [PubMed] [Google Scholar]
- 66.Del Prete, G., M. De Carli, F. Almerigogna, M.G. Giudizi, R. Biagiotti, and S. Romagnani. 1993. Human IL-10 is produced by both type 1 helper (Th1) and type 2 helper (Th2) T cell clones and inhibits their antigen-specific proliferation and cytokine production. J. Immunol. 150:353–360. [PubMed] [Google Scholar]
- 67.Noma, T., H. Nakakubo, M. Sugita, S. Kumagai, M. Maeda, A. Shimizu, and T. Honjo. 1989. Expression of different combinations of interleukins by human T cell leukemic cell lines that are clonally related. J. Exp. Med. 169:1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paliard, X., R. de Waal Malefijt, H. Yssel, D. Blanchard, I. Chretien, J. Abrams, J. de Vries, and H. Spits. 1988. Simultaneous production of IL-2, IL-4, and IFN-gamma by activated human CD4+ and CD8+ T cell clones. J. Immunol. 141:849–855. [PubMed] [Google Scholar]
- 69.Pirmez, C., M. Yamamura, K. Uyemura, M. Paes-Oliveira, F. Conceicao-Silva, and R.L. Modlin. 1993. Cytokine patterns in the pathogenesis of human leishmaniasis. J. Clin. Invest. 91:1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romagnani, S. 1991. Human TH1 and TH2 subsets: doubt no more. Immunol. Today. 12:256–257. [DOI] [PubMed] [Google Scholar]
- 71.Messi, M., I. Giacchetto, K. Nagata, A. Lanzavecchia, G. Natoli, and F. Sallusto. 2003. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat. Immunol. 4:78–86. [DOI] [PubMed] [Google Scholar]
- 72.Murphy, E., K. Shibuya, N. Hosken, P. Openshaw, V. Maino, K. Davis, K. Murphy, and A. O'Garra. 1996. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J. Exp. Med. 183:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Overbergh, L., D. Valckx, M. Waer, and C. Mathieu. 1999. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine. 11:305–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.