Abstract
TCRαβ signaling is crucial for the maturation of CD4 and CD8 T cells, but the role of the Notch signaling pathway in this process is poorly understood. Genes encoding Presenilin (PS) 1/2 were deleted to prevent activation of the multiple Notch receptors expressed by developing thymocytes. PS1/2 knockout thymocyte precursors inefficiently generate CD4 T cells, a phenotype that is most pronounced when thymocytes bear a single major histocompatibility complex (MHC) class II–restricted T cell receptor (TCR). Diminished T cell production correlated with evidence of impaired TCR signaling, and could be rescued by manipulations that enhance MHC recognition. Although Notch appears to directly regulate binary fate decisions in many systems, these findings suggest a model in which PS-dependent Notch signaling influences positive selection and the development of αβ T cells by modifying TCR signal transduction.
Upon recognition of MHC ligands on thymic stroma, TCRαβ signaling in CD4+CD8+ (double-positive [DP]) thymocytes mediates positive selection and the development of CD4 and CD8 αβ T cells. There is broad support for quantitative models in which the strength/duration of TCR signals determines the fate of a DP thymocyte. Accordingly, the strongest TCR signals induce negative selection by deletion; intermediate TCR signals promote positive selection and maturation to the single-positive (SP), CD4+CD8− or CD4−CD8+, stage; and weak or no TCR signals result in cell death. The quantity of the TCR signal has also been proposed to be a primary determinant in CD4 and CD8 lineage commitment. Among DP thymocytes receiving TCR signals within the range for positive selection, those receiving stronger/prolonged signals are biased toward the CD4 fate, whereas those experiencing weaker/transient signals adopt the CD8 fate (1-3). A provocative and lingering topic in this field is how quantitatively different TCR signals in DP thymocytes are translated intracellularly to specify these T cell fates.
The Notch signaling pathway is another important regulator of cell fate in thymocytes. Notch is essential for commitment of lymphoid precursors to the T cell lineage. Notch signaling is required in double-negative (DN) CD4−CD8− thymocytes for maturation through the DN1–DN4 stages, where it has been associated with such critical functions as TCRβ rearrangement, pre-TCR signaling, and αβ/γδ T lineage commitment (4-12). Notch has also been implicated in the subsequent CD4 versus CD8 lineage decision that is initiated in DP thymocytes (13–15). Collectively, these data suggest that recurrent/continuous Notch signaling may be required throughout thymocyte development.
Nevertheless, several experimental approaches have yielded conflicting results on the role of Notch signaling in the late stages of αβ T cell development. Development of SP thymocytes is normal in mice deficient for individual Notch family members (16–19). Inhibiting Notch signaling either by conditional deletion of the transcriptional regulator CBF1/Su(H)/Lag-1 (CSL; also called RBPJκ) or expression of a dn form of the transcriptional coactivator Mastermind (dnMAML), were also reported to have no effect on CD4/CD8 T cell fate (7, 20). Likewise, overexpression of Numb, which is an antagonist of Notch, has no obvious effect on development (21).
In contrast to these negative results, other studies imply that Notch does play a role in CD4 and CD8 T cell development. When pharmacological inhibitors were added to fetal thymic organ cultures to block Notch activation, the authors of two independent studies concluded that Notch activity promotes CD8 development (22, 23). Late thymocyte development is also altered in several transgenic (Tg) mouse lines expressing the constitutively active Notch intracellular domain (NICD) (13–15, 24, 25). If mice are analyzed before the appearance of tumors, development of SP thymocytes is MHC dependent, and NICD favors development of the CD8 over the CD4 lineage. Because NICD redirected some MHC class II–specific cells to the CD8 lineage, it was proposed that Notch plays a role in CD4/CD8 lineage commitment. Consequently, the conflicting results obtained from gain- and loss-of-function Notch studies, along with the potential for redundancy between Notch family members and examples of CSL-independent Notch signaling (26), leave open the question of whether endogenous Notch functions in CD4 and CD8 T cell development and lineage commitment.
The evidence that both TCR and Notch play a role in the development of SP thymocytes raises the issue of whether these two signaling pathways are functionally linked. Studies of mature T cells claiming that TCR stimulation promotes the cleavage of Notch are relevant (27–29). Conversely, there is evidence that Notch signaling modulates TCR signaling in mature T cells (29–31). Another suggestion that Notch and TCR signals may be integrated comes from results showing that Notch coclusters with the TCR in antigen-stimulated thymocytes (32). In early thymocytes, Notch and pre-TCR signals are both required to mediate the DN–DP transition (4), and in DP thymocytes, a NICD Tg enhances the effects of TCR stimulation in regulating gene expression (33). In contrast, high levels of NICD were interpreted to inhibit TCR signals in a study where DP to SP thymocyte development was blocked completely (34). Thus, there is evidence for TCR signaling regulating Notch activation, Notch activity regulating TCR signaling, and evidence that these pathways are parallel but linked. At the very least, these results suggest some level of cross-talk, which could involve several distinct mechanisms.
To further investigate these issues, we developed a new approach for inhibiting endogenous Notch signaling in thymocytes that avoids the problems of embryonic lethality, redundancy, and Notch-induced malignancies that plagued many of the earlier studies. The best-characterized pathway of how Notch signaling regulates gene expression involves the conversion of a transcriptional repressor complex to a transcriptional activating complex. Upon ligand engagement, Notch receptors undergo two proteolytic cleavages, the second involving a presenilin (PS)-containing complex with γ-secretase activity that releases NICD. NICD translocates from the membrane to the nucleus, where it binds to CSL, displacing corepressors and recruiting coactivators. Because PSs are required for the activation of all four mammalian forms of Notch, we generated mice with deletions of both Presenilin1 (PS1) and PS2 genes, the only genes encoding PS in the mouse genome. To target Notch inactivation specifically to developing T cells, we introduced Cd4Cre to mediate PS gene deletion in a tissue- and stage-specific manner. We find that rather than directing CD4/CD8 T lineage cell commitment, the data demonstrate that PS-dependent Notch signaling influences αβ T cell maturation by modifying TCR signal strength in DP thymocytes.
RESULTS
Generation of conditional PS1/2KO mice
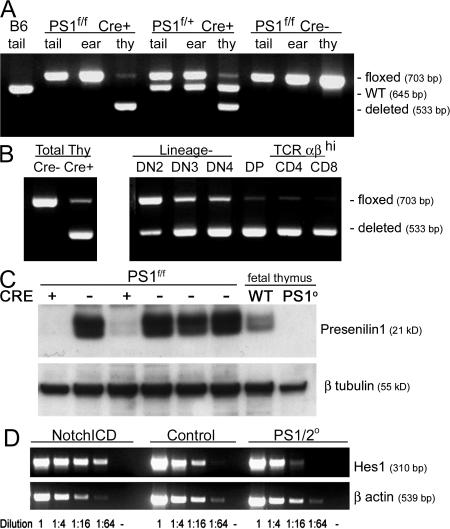
PS1 and PS2 are expressed by thymocytes (Fig. 1 C and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070550/DC1). PS2-null mice have normal lymphocyte development (Fig. 2 and not depicted). Although PS1-null mutant mice die near the time of birth, analyses of hematopoietic chimeras made with fetal liver of the mutant indicated that lymphocyte development was largely normal (unpublished data). To avoid the embryonic lethality associated with gene deletion of both PS1 and PS2 (35, 36), we targeted deletion of a floxed allele of the PS1 gene (37) specifically to developing T cells by expressing a Cd4Cre Tg (38) in the PS2-null background. With this conditional deletion, the deleted PS1 gene is detectable only in T lineage cells (Fig. 1 A).
Figure 1.
Efficient deletion of the floxed PS1 allele by Cd4Cre in developing thymocytes. (A) PCR amplification of PS1 from the ear, tail, or thymus discriminates between the undeleted floxed allele (703 bp), the deleted allele (533 bp), and the endogenous (WT) PS1 gene (645 bp). A representative experiment assaying deletion in homozygous (lanes 2–4) or heterozygous (lane 5–7) PS1-floxed mice is shown. No deletion is seen in the absence of Cre (lanes 8–10). (B) Conditional PS1 gene deletion in total thymocytes (left) was staged by PCR analysis of sorted thymocytes from PS1 f/f Cre+ mice (right). (C) Western blot analysis for PS1 protein in lysates of thymocytes from six individual littermates bearing floxed PS1 with or without Cre expression. Lysates from fetal thymus of PS1+ and PS1−/− littermates are included as antisera specificity controls. (D) Semiquantitative RT-PCR analysis for Hes1 expression performed using total RNA isolated from thymocytes of transgenic pLck- NICD, control, or PS1/2KO mice. Template cDNA was serially diluted as indicated. A reaction with no cDNA template added (-) serves as a negative control.
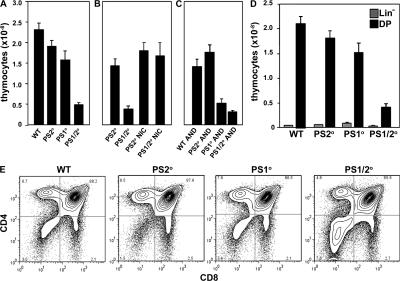
Figure 2.
Thymus cellularity is decreased in the absence of PSs. (A) Mean thymus cellularity of WT, PS2 KO, PS1 KO, and PS1/2KO mice. (B) Mean thymus cellularity of control and PS1/2KO with or without expression of transgenic NotchIC (NICD). (C) Mean thymus cellularity of H-2b WT, PS2 KO, PS1 KO, and PS1/2KO mice bearing transgenic AND TCR on a H-2b RAG2° background. (D) Mean number of lineage negative (gray bars) and CD4+CD8+ DP (black bars) thymocytes isolated from WT, PS2 KO, PS1 KO, and PS1/2KO mice. (E) Total thymocytes isolated from WT, PS2 KO, PS1 KO, and PS1/2KO littermates analyzed by flow cytometry for expression of CD4 and CD8. Error bars represent the SEM. n = 9–48 mice.
A faint band representing the floxed (f) PS1 allele was detectable in PCR analyses of DNA isolated from thymocytes of mice with PS1f/fCre+ and PS1f/+Cre+ genotypes (Fig. 1 A). To determine the precise timing of PS1 gene deletion, DNA was isolated from sorted lineage-negative (Lin−), DP, or TCRαβhi CD4 or CD8 SP thymocytes. PS1 deletion was detectable as early as the DN2 (CD44+CD25+) stage (Fig. 1 B), and the floxed PS1 allele was virtually undetectable by the DP stage and in more mature TCRαβhi CD4 and CD8 SP thymocytes. PS1 protein was nearly absent in thymocytes of Cd4Cre+PS1f/f mice (Fig. 1 C). These data indicate that both PS1 genes and PS1 protein are efficiently eliminated in thymocytes of PS1/2 conditional KO mice.
Because Notch activation requires PS-dependent cleavage, deleting PS genes should inhibit all Notch signaling. Expression levels of a Notch target gene, Hes1, were analyzed by semiquantitative RT-PCR (Fig. 1 D). Compared with wild-type, Hes1 was elevated in thymocytes expressing constitutively active Notch1ICD, and decreased in PS1/2KO thymocytes, which was an indication that Notch signaling was impaired.
Decreased thymus cellularity in the absence of PS
To assess the impact of PS deficiency on thymopoiesis, thymocyte numbers were analyzed. Thymus cellularity is normal in mice deficient for PS2 (Fig. 2 A), but is reduced in mice deficient for PS1 (P = 0.01). Thymus cellularity is further reduced in mice lacking both PS1 and PS2 (P < 0.0001). Thymus cellularity is similarly reduced in chimeric mice made by reconstituting irradiated recipients with PS1/2KO BM, confirming that decreased cellularity is a cell-intrinsic defect of PS1/2KO thymocytes (unpublished data). Moreover, thymus cellularity is unaffected in mice expressing a Cd4Cre Tg alone, with no floxed allele (unpublished data). Because NICD is constitutively active in the absence of PS, we expressed a NICD Tg in PS1/2KO thymocytes. Thymus cellularity is restored to control levels by NICD (Fig. 2 B), supporting the hypothesis that decreased thymus cellularity in PS1/2KO mice is caused by an absence of Notch cleavage.
Although thymocyte numbers are reduced, PS deficiency does not cause a complete block in development, but rather alters the distribution of thymocyte subsets, such that the proportion of DN to DP thymocytes is increased (Fig. 2 E). Because no substantial differences between the wild-type and PS2-deficient mice were observed (Fig. 2 and not depicted), all subsequent analyses were performed using PS2KO littermates as controls for conditional PS1/2KO mice.
The number of Lin− thymocytes is unchanged, whereas DP thymocyte number is dramatically reduced (Fig. 2 D), suggesting that in the absence of PS, fewer thymocytes mature from the DN to the DP stage. A similar phenotype was recently observed in mice expressing an inducible dnMAML (12), and these in vivo results corroborate the in vitro results of Ciofani et al. (4), who find that both Notch and pre-TCR signals are absolutely required to mediate an efficient DN–DP transition. Thus, we conclude that with Cd4Cre-mediated PS gene deletion, thymocytes that lose PS before the DN3 stage fail to progress to the DP stage, whereas the DPs that are present must have lost PS after β selection was initiated.
CD4 thymocytes are generated inefficiently in the absence of PS
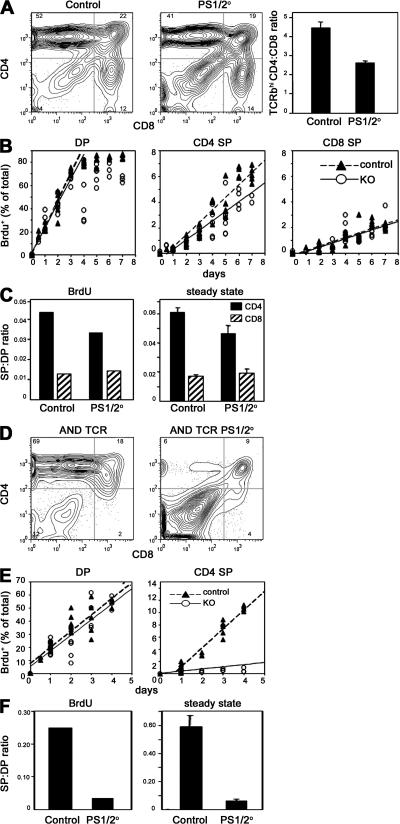
To address the role of PS in positive selection and the development of SP thymocytes, we analyzed TCRαβhi thymocytes. Surprisingly, there are proportionately fewer CD4, and more CD8, SP thymocytes in PS1/2KO mice, resulting in a decreased ratio of TCRαβhi CD4/CD8 thymocytes compared with that of controls (P < 0.0001; Fig. 3 A). This phenotype is also evident in 4-d-old pups (unpublished data), suggesting that the differences are not caused by accumulation of mature thymocytes.
Figure 3.
CD4 SP thymocytes are inefficiently generated in the absence of PSs. (A) Representative FACS plots analyzing the expression of CD4 and CD8 among the TCRβhi thymocytes from control and PS1/2KO mice. The bar graph shows the mean ratio of TCRβhi CD4 to CD8 thymocytes, which was calculated from cell counts and flow cytometric analysis of individual mice. Error bars represent the SEM. n = 37–40 mice. (B) Control and PS1/2KO thymocytes were labeled continuously with BrdU, as described in the Materials and methods. The kinetics of BrdU incorporation within the DP (CD4+CD8+) or TCRhi CD4 and CD8 SP thymocytes is plotted as a percentage of total thymocytes. Closed triangles and dotted lines represent control thymocytes. Open circles and solid lines represent PS1/2KO thymocytes. The slopes of the lines did not differ significantly between DP or CD8 SP thymocytes from control or KO mice, but were significantly different for control and KO CD4 SP thymocytes (P = 0.01). (C) Efficiency of generating mature CD4 SP (filled bar) or CD8 SP (hatched bar) thymocytes in control or PS1/2KO mice provided with continuous BrdU was calculated by dividing the rate of mature SP generated per day by the rate of immature DP thymocytes generated per day (left). Efficiency of generating mature CD4 SP (filled bar) or CD8 SP (hatched bar) in the steady state was estimated by determining the ratio of mature SP to immature DP thymocytes isolated from individual mice (right). Error bars represent the SEM. n = 37–40 mice. (D) Thymocytes from H-2b control and PS1/2KO mice bearing AND TCR analyzed for coexpression of CD4 and CD8 and gated for TCRVα11hi. (E) H-2b control (closed triangles and dashed lines) and PS1/2KO thymocytes (open circles and solid lines) bearing AND TCR labeled with continuous BrdU as described in B. Data points for BrdU+ CD4+CD8+ DP or TCRhi CD4 SP cells are shown as a percentage of total thymocytes plotted against labeling time. The slopes of the lines did not differ significantly between control and KO DP thymocytes, but were significantly different for control and KO CD4 SP thymocytes (P < 0.0001). (F) Efficiency of generating mature CD4 SP cells in H-2b control and PS1/2KO mice bearing AND TCR provided with continuous BrdU was estimated from the ratio of CD4 SP to DP thymocytes, as described in C (left). Efficiency of generating mature CD4 SP in steady state was estimated from the ratio of CD4 SP to DP thymocytes as described in C (P < 0.0001; right). Error bars represent the SEM. n = 36–46 mice.
The loss of DP thymocytes associated with PS deficiency leads to decreased numbers of both CD4 and CD8 SP cells, preventing a direct comparison with SP cell numbers in control animals. To circumvent this problem, we assessed the efficiency of generating SP thymocytes from DP precursors in two ways. In one approach, mice were exposed to in vivo continuous BrdU labeling (39, 40). The percentage of BrdU+ cells for each subset was determined (Fig. 3 B). If the slope of the line representing BrdU uptake over the first several days is used to estimate the rate of DP thymocyte generation, control animals generated 3.2 × 107 DP cells per day, whereas PS1/2KO animals generated only 5.5 × 106 DP cells per day. The rate of SP thymocyte generation was similarly estimated, revealing that control animals generated 1.4 × 106 and 4.2 × 105 CD4 and CD8 SP cells per day, respectively, resulting in a CD4/CD8 ratio of 3.3:1. In contrast, PS1/2KO mice generated only 1.8 × 105 and 7.8 × 104 mature CD4 and CD8 SP cells per day, respectively, resulting in a CD4/CD8 ratio of only 2.3:1. Dividing the rate of CD4 generation by the rate of DP generation provides an estimate of the rate of conversion of DP to SP (39, 40). From this calculation, the efficiency of DP to CD4 SP thymocyte conversion is ∼4.3% for the controls, but drops by 25% to only 3.3% in the absence of PS1/2 (Fig. 3 C). In contrast, CD8 SP cells appear to be generated as efficiently in PS1/2KO as in control mice, at 1.3 and 1.4%, respectively. In a second approach, an estimate of the rate of conversion was made based on the size of thymocyte subsets in the steady-state thymus by simply dividing the number of TCRαβhi SP thymocytes by the number of DP thymocytes. Although the absolute values differ slightly from those determined in the BrdU-labeling experiments, the same trends are observed (Fig. 3 C). The efficiency of generating CD4 SP from DP precursors is lower in PS1/2KO mice than in controls (P = 0.03), whereas the generation of CD8 SP is similar. Thus, by either analysis, CD4 SP thymocytes are inefficiently generated in the absence of PS in mice with a diverse TCR repertoire.
Decreased generation of SP T cells in PS1/2KO thymocytes expressing TCRαβ Tg
Although impaired CD4 SP generation in PS1/2KO mice is a consistent and statistically significant finding, the decrease is fairly subtle. Even so, alterations in cell fates associated with changes in TCR signaling can be masked at the population level by the selection of a different TCR repertoire (41–43). To assess the affect of PS deficiency on CD4 development in mice bearing a single TCRαβ, we generated RAG2° PS1/2KO mice expressing MHC class II–restricted AND TCR. Similar to mice with the diverse TCR repertoire, thymus cellularity was decreased in PS1KO (P = 0.009) mice and severely reduced in PS1/2KO mice (P < 0.0001; Fig. 2 C). Although CD4 SP thymocytes are generated extremely well in control H-2b AND TCR mice, very few TCRhi CD4 SP cells are generated in PS1/2KO littermates (Fig. 3 D). Continuous BrdU labeling studies indicated that control AND TCR mice generated 2.6 × 107 DP and 6.4 × 106 CD4 SP thymocytes per day, respectively, whereas PS1/2KO AND TCR transgenic animals generated only 2.9 × 106 DP and 9.9 × 104 CD4 SP cells per day, respectively (Fig. 3 E). In control AND TCR mice, 25% of DP cells are positively selected to become CD4 SP T cells (Fig. 3 F). In PS1/2KO AND TCR mice, the efficiency of DP to CD4 SP conversion drops precipitously to only 3%. The same trend is observed when the rate of conversion was calculated based on the CD4/CD8 subsets in the steady-state thymus (Fig. 3 F). Similar data were obtained with mice expressing a 5CC7 TCR, although the defect in generating CD4 SP thymocytes is less severe (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070550/DC1). The difference is most likely explained by the proposal that 5CC7 TCR has a higher avidity for selecting ligand than AND TCR (44). These data suggest that PS-dependent Notch signaling may be most important for positive selection under conditions where selecting ligand and/or TCR affinity are limiting.
In PS1/2KO mice with a diverse TCR repertoire, no differences in the efficiency of CD8 generation are detected (Fig. 3). However, when the efficiency of DP to CD8 SP conversion for thymocytes bearing a single class I–restricted TCRαβ is analyzed, it is revealed that the generation of CD8 lineage cells is also impaired, albeit less so than CD4 lineage cells (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070550/DC1). By focusing on the selection of a single TCR, either class I or class II restricted, our analyses reveal that development of both CD4 and CD8 lineage cells can be impaired in the absence of PS.
Attenuated TCR signaling in PS1/2KO thymocytes
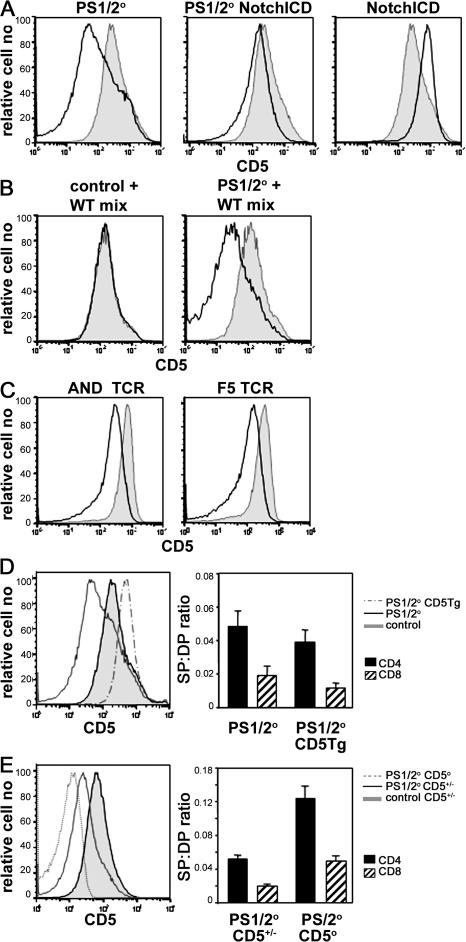
We considered the possibility that defective TCR signaling was responsible for the impaired CD4 T cell development in PS1/2KO mice. CD5 expression has been correlated with TCR signal strength (45). Despite similar levels of TCR, the levels of CD5 expressed by DP thymocytes were dramatically reduced in PS1/2KO mice compared with control mice (Fig. 4 A). CD5 levels were increased in DP cells expressing NICD, and CD5 expression by PS1/2KO DP was restored to wild-type levels by expression of NICD. In mixed BM chimeras, CD5 levels were markedly lower on PS1/2KO-derived DP thymocytes than on wild-type–derived DP cells developing in the same host (Fig. 4 B), but were identical for control- and wild-type–derived DP thymocytes developing in the same host (Fig. 4 B), confirming that decreased CD5 expression is a cell autonomous defect of PS1/2KO DP thymocytes. To exclude the possibility that changes in TCR repertoire were responsible for the changes in CD5 expression, we compared CD5 expression on DP thymocytes expressing a fixed TCRαβ. As with the diverse TCR repertoire, the level of CD5 was significantly reduced on DP thymocytes of class II–restricted AND TCR and 5CC7 TCR and class I–restricted F5 TCR Tg mice (Fig. 4 C and Fig. 6) when PSs were absent. Collectively, these data suggested that CD5 expression by DP thymocytes is regulated by PS.
Figure 4.
Abnormally low CD5 expression by DP thymocytes in the absence of PSs. (A) Analysis for expression of CD5 on CD4+CD8+ DP thymocytes isolated from PS1/2KO (left, open histogram), PS1/2KO NotchICD (middle, open histogram), and NotchICD (right, open histogram) littermates. Analysis of a control littermate (shaded histogram) is overlaid for comparison. (B) Analysis for expression of CD5 on CD4+CD8+ DP thymocytes in mixed radiation chimeras that were prepared as described in Materials and methods. Histograms representing CD5 levels of wild-type B6 CD45.1+-derived DP thymocytes (shaded histogram) overlaid with CD45.2+-derived (open histogram) control (left) or PS1/2KO (right) DP thymocytes that developed in the same host. (C) Analysis for CD5 expression on DP thymocytes isolated from H-2b control (shaded histogram) and PS1/2KO (open histogram) littermates bearing AND (left) or F5 (right) TCR. (D) Analysis for expression of CD5 on DP thymocytes isolated from PS1/2KO littermates, with (dashed line) or without (solid line) a CD5 Tg. CD5 expressed on DP thymocytes isolated from a control littermate (shaded histogram) is overlaid for reference. An estimate of the mean efficiency of generating mature CD4 SP (filled bar) and CD8 SP (hatched bar) thymocytes in the steady state was estimated as in Fig. 3 C. Error bars represent the SEM. n =13–16 mice. (E) DP thymocytes isolated from CD5+/− (solid line) or CD5−/− (dotted line) PS1/2KO littermates and analyzed for expression of CD5. CD5 expression on CD4+CD8+ DP thymocytes isolated from a control CD5+/− littermate (shaded histogram) is overlaid for reference. An estimate of the mean efficiency of generating mature CD4 SP (filled bar) and CD8 SP (hatched bar) thymocytes in the steady state was estimated as in Fig. 3 C. Error bars represent the SEM. n = 15–20 mice.
Figure 6.
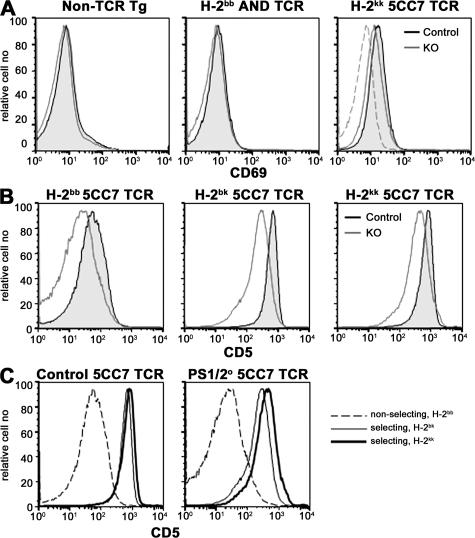
PS-deficient DP thymocytes respond to selecting MHC by up-regulating CD5 and CD69. (A) Analysis for CD69 expression on CD4+CD8+ DP thymocytes isolated from control (shaded histogram) and PS1/2KO (open histogram) mice with either a diverse TCR repertoire (left), an AND TCR (middle), or a 5CC7 TCR (right) in a selecting MHC background. CD69 expressed on DP thymocytes isolated from control mice bearing 5CC7 TCR on the nonselecting H-2b background (dashed line) are overlaid for reference (right). (B) Analysis for expression of CD5 on positive DP thymocytes isolated from control (shaded histogram) and PS1/2KO (open) mice bearing 5CC7 TCR on the nonselecting H-2b, or selecting H-2bk or H-2kk background. (C) Same data as in B, replotted with control (left) or PS1/2KO (right) thymocytes of nonselecting H-2b (dashed lines) or selecting H-2bk (thin line) or H-2kk (thick line) background, overlaid for comparison.
It was not clear whether the alterations in CD5 expression caused or reflected the changes in thymic selection observed with PS deficiency. Because CD5 is a negative regulator of TCR signaling (41–43), its decreased expression would be expected to enhance TCR signal strength, and perhaps even promote negative selection. On the other hand, PS deficiency could reduce TCR signal strength at the DP stage, which could be reflected in altered CD5 levels as a consequence. To distinguish between these possibilities, we examined the impact of forced CD5 expression on the PS1/2KO phenotype. If Notch controlled CD5 expression, and CD5 modulation was directly responsible for abnormal CD4 SP thymocyte development in PS1/2KO mice, then restoration of CD5 levels should correct this defect. To the contrary, a CD5 Tg increased CD5 expression, but exacerbated the PS1/2KO phenotype (Fig. 4 D and not depicted). With the CD5 Tg, thymus cellularity was further decreased and the generation of both CD4 and CD8 SP thymocytes was even more impaired. Conversely, removing CD5 entirely from PS1/2KO thymocytes results in increased thymus cellularity and improved generation of SP thymocytes (Fig. 4 E and not depicted). These data are consistent with the notion that reduced expression of CD5 is a reflection of diminished in vivo TCR signal strength in PS1/2KO DP thymocytes.
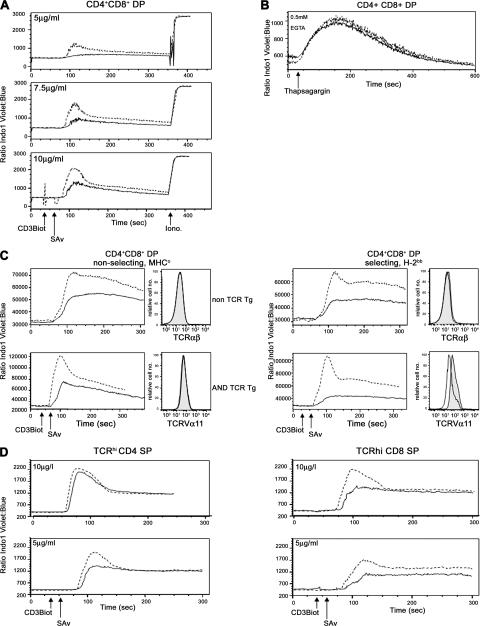
The low levels of CD5 on DP thymocytes and the exacerbation of the PS1/2KO phenotype by restoration of CD5 expression suggested that TCR signaling was defective in the absence of PS. Because the magnitude of calcium/calcineurin signals plays a crucial role in TCR signaling and positive selection (46, 47), Ca2+ mobilization was analyzed in PS1/2KO thymocytes in response to TCR stimulation in vitro. Although the calcium ionophore ionomycin elicited equivalent responses in control and PS1/2KO thymocytes, the magnitude of Ca2+ flux was attenuated in PS1/2KO DP thymocytes at every dose of anti-CD3 tested (Fig. 5 A).
Figure 5.
Calcium flux in response to TCR cross-linking is reduced in PS-deficient DP thymocytes. (A) Mean of calcium flux measurements in CD4+CD8+ DP thymocytes isolated from control (dashed line) and PS1/2KO (solid line) after stimulation with concentrations of anti-CD3 indicated in the insets. (B) Calcium fluxes of DP thymocytes isolated from control (dashed line) and PS1/2KO (solid line) littermates in Ca-free media after the addition of thapsigargin. (C) Analysis for Ca2+ fluxes in thymocytes generated from BM chimeras made in MHC-deficient (left) or MHC-expressing (right) hosts. Calcium fluxes in PS2KO (dashed lines) or PS1/2KO (solid lines) DP thymocytes are compared in either mice with a diverse TCR repertoire (top) or an AND TCR (bottom). Histograms beside each calcium trace plot the levels of TCR expression by DP thymocytes derived from control PS2KO (shaded histogram) or PS1/2KO (open histogram) BM. (D) Analysis for Ca2+ flux as in A by TCRhi CD4+ or CD8+ SP thymocytes isolated from control (dotted line) and PS1/2KO (solid line) littermates.
Although controversial, PS has been implicated to play a role in maintaining intracellular Ca2+ stores in some systems/cell types. To the contrary, treatment of control and PS1/2KO thymocytes with the sarcoendoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin, in the absence of extracellular Ca2+, demonstrates that PS1/2KO DP thymocytes have normal intracellular Ca2+ stores (Fig. 5 B).
We considered the possibility that PS1/2KO DP thymocytes flux Ca2+ poorly because they are less mature than DP isolated from control littermates. To analyze a more synchronized population, we examined DP thymocytes of mice developing in the absence of selecting MHC (Fig. 5 C and Fig. S1). Because both control and PS1/2KO DP thymocytes are developmentally arrested at a comparable stage in the nonselecting background, levels of TCR expression are equivalent. Nevertheless, Ca2+ flux is markedly reduced in the PS1/2KO cells, indicating that the TCR signaling defect is intrinsic to PS1/2KO thymocytes and that the defect is initiated before positive selection.
As controls for the MHC° hosts, wild-type MHC+ mice were also irradiated and reconstituted with BM from control or PS1/2KO mice (Fig. 5 C). As observed with intact mice, PS1/2KO DP thymocytes with a diverse TCR repertoire have normal levels of TCR expression, but reduced Ca2+ flux, in response to TCR cross-linking, confirming that the signaling defect is intrinsic to DP thymocytes. In contrast to mice with a diverse TCR repertoire, however, PS1/2KO DP thymocytes express low levels of AND TCR (Fig. 5 C). Because TCR up-regulation is a hallmark of positive selection, these lower levels of TCR, coupled with the reduced Ca2+ response and paucity of mature CD4 SP thymocytes, suggest that positive selection is severely impaired in the absence of PS.
Overexpression of NICD leads to increased expression of Bcl-2, which confers resistance to apoptosis (48, 49). We assessed cell death to exclude the possibility that the TCR signaling defect in PS1/2KO thymocytes was related to reduced cell survival. As assayed by loss of mitochondrial potential or by Annexin V staining, there was no difference in the percentage of apoptotic cells ex vivo (unpublished data).
To determine if the signaling defect was maintained in mature thymocytes, the ability of SP thymocytes to flux Ca2+ in response to TCR cross-linking was analyzed (Fig. 5 D). CD4 SP thymocytes of control and PS1/2KO mice flux Ca2+ equally well at higher doses of TCR cross-linking; however, when lower doses of anti-CD3 were used, the magnitude of Ca2+ flux was attenuated in CD4 SP PS1/2KO cells. This phenotype was even more pronounced in CD8 SP thymocytes than CD4 SP thymocytes. These data indicate that PSs influence TCR signaling in both MHC class I– and class II–restricted T cells when antigen is limiting. These data are consistent with the observation that the ability of Notch signaling to augment the responsiveness of mature T cells is most prominent when T cells are stimulated with low-dose anti-CD3 in the absence of costimulation (27).
Responses of PS1/2KO thymocytes to TCR stimulation
Control and PS1/2KO DP thymocytes express comparable levels of CD69 ex vivo, indicating that TCR signals transduced by positive selection in PS1/2KO thymocytes are sufficient to induce CD69 expression (Fig. 6 A). This was not surprising because CD69 is an early activation marker that can be induced by low levels of TCR signaling (50). Moreover, control and PS1/2KO up-regulate CD69 equally well after TCR stimulation (unpublished data).
Although the response is muted, PS1/2KO DP thymocytes flux Ca2+ in a dose-dependent manner in response to anti-CD3, indicating that they do sense differences in TCR signal strength in vitro. To determine if this was true in vivo, we measured ex vivo CD5 expression on DP thymocytes isolated from 5CC7 TCR mice expressing increasing amounts of the selecting MHC. Because 5CC7 TCR is selected by class II Ek, but not by class II, of H-2b mice, CD5 expression was analyzed on DP thymocytes in nonselecting (H-2bb) or selecting (H-2bk or H-2kk) mice. CD5 expression is up-regulated on DP thymocytes of 5CC7 TCR mice in response to selecting versus nonselecting MHC, irrespective of the PS deficiency (Fig. 6 C). Moreover, for both PS1/2KO and control DP thymocytes, CD5 levels correlate with increasing amounts of selecting MHC (H-2bb < H-2bk < H-2kk). However, within each MHC haplotype, PS1/2KO DP cells express less CD5 than controls (Fig. 6 B). These in vivo results are analogous to the in vitro Ca2+ response, demonstrating that PS1/2KO DP thymocytes do not respond as well as controls across a range of selecting MHCs.
Higher affinity MHC ligand improves selection of PS1/2KO SP thymocytes
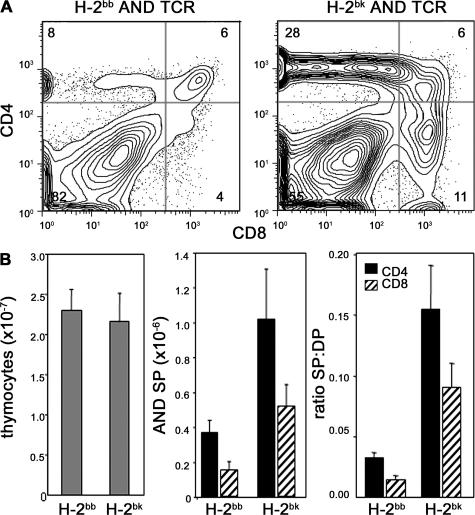
Although PS1/2KO DP thymocytes are somewhat compromised, they are capable of responding to TCR stimuli as indicated by their ability to up-regulate CD69 and CD5 in vivo, and to mobilize Ca2+ and induce CD69 in response to anti-CD3 in vitro. If attenuated TCR signaling in PS1/2KO DP cells were responsible for the impaired development of CD4 and CD8 SP thymocytes, then SP generation should be restored with higher affinity and/or density of selecting MHC ligands. To determine if higher affinity TCR–MHC interactions could improve positive selection of PS1/2KO thymocytes, we compared selection of AND TCR–bearing thymocytes in mice expressing MHC class II Ek, to those expressing Ab. Although both Ek and Ab mediate positive selection of the AND TCR, Ek is better recognized than Ab, and can even promote some negative selection (51–53). Because thymus cellularity was similar in PS1/2KO H-2bb and H-2bk AND TCR mice, both the number and the efficiency of SP thymocyte generation could be compared (Fig. 7). Indeed, positive selection of PS1/2KO SP thymocytes is greater by H-2bk than by H-2bb (P = 0.01 and 0.03 for CD4 SP and CD8 SP, respectively). Similarly, positive selection of PS1/2KO thymocytes bearing 5CC7 TCR is markedly better on H-2kk than on H-2bk (Fig. S1). Thus, increasing either the TCR affinity or the density of selecting MHC can compensate for the attenuated TCR signaling that diminishes positive selection in the absence of PS.
Figure 7.
Inefficient generation of CD4 SP thymocytes in PS-deficient mice is improved by increasing affinity of selecting MHC ligand. (A) Thymocytes of PS1/2KO mice bearing AND TCR on a H-2bb or H-2bk background, gated for TCRVα11hi and analyzed for expression of CD4 and CD8. (B) PS1/2KO mice bearing AND TCR on a H-2bb or H-2bk background analyzed for mean thymus cellularity (left), mean number of TCRVα11hi CD4 and CD8 SP thymocytes (middle), and the mean efficiency of generating mature CD4 SP and CD8 SP thymocytes in the steady state (right), calculated as in Fig. 3 C. Error bars represent the SEM. n =16–26 mice.
DISCUSSION
Several studies suggested that Notch influences the CD4/CD8 lineage decision, favoring the CD8 over the CD4 fate. These results fit with evidence from many species, indicating that Notch signaling directs binary fate decisions. On the other hand, attempts to interfere with endogenous Notch have had no obvious effect on thymic selection or development of mature αβ lineage T cells. Our studies reveal that PS-dependent Notch signaling does, in fact, play a role in generating mature thymocytes, but rather than directing lineage commitment, Notch mediates its effects by modifying TCR signal transduction. Importantly, these results indicate that TCR and Notch signaling pathways are functionally linked in the selection and maturation of αβ T cells.
Our approach was to generate mice with conditionally deleted PS genes as a means of inhibiting all endogenous Notch signaling in developing T cells. The generation of CD4 SP thymocytes from DP precursors was inefficient in mice with a diverse TCR repertoire, and was severely compromised in mice expressing a single MHC class II–restricted TCR. Defective CD4 T cell development was associated with impaired TCR signaling at the DP stage, as indicated by a reduced Ca2+ response to TCR stimulation in vitro and decreased CD5 up-regulation in response to selecting MHC in vivo. Moreover, the fact that CD5 expression and the generation of CD4 SP thymocytes could be improved by higher affinity or density of selecting MHC ligand demonstrated that defective CD4 development was a consequence of poor positive selection in PS1/2KO thymocytes. The number and phenotype of DP was corrected by expression of a constitutively active form of Notch, indicating that the phenotypes observed in PS1/2KO thymocytes were caused by a loss of Notch signaling. Collectively, our findings suggest that under physiological conditions, PS-dependent Notch signaling influences positive selection and αβ T cell maturation by potentiating TCR signaling in DP thymocytes.
Although the adverse effect of PS-deficiency on production of CD4 SP was obvious, the effect on CD8 SP thymocytes was evident only in mice expressing a single MHC class I–restricted TCR. Even though CD8 SP generation appeared normal with a diverse TCR repertoire, the Ca2+ response to TCR stimulation of these cells was attenuated and CD5 expression was reduced in DP thymocytes bearing a MHC class I–restricted TCR, which are both indicators of impaired CD8 T cell development. Notably, a differential affect on the development of CD4 versus CD8 SP thymocytes is also observed in other mice with deficiencies in TCR signaling (54–57). Given that TCR signaling is required for positive selection of both CD4 and CD8 T cells, the question arises as to why reduced TCR signaling in DP thymocytes has a greater impact on the generation of CD4 cells? The difference is most easily explained by a quantitative signaling model in which higher TCR signals are required for CD4 than for CD8 T cell commitment (1-3), such that selection of CD4 T cells would be most affected if TCR signaling was attenuated. The finding that higher density or affinity MHC ligands rescue development of class II–restricted CD4 SP thymocytes in PS1/2KO mice is also explained by the model, as is the observation that selection of thymocytes with 5CC7 TCR is less severely impaired than those with AND TCR in PS1/2KO mice.
How might PS affect TCR signaling in DP thymocytes? It is well established that PSs are absolutely required for Notch activation, and that Notch signaling is important for the DN–DP transition. The requirement for Notch activation in DN3 thymocytes is absolute; thymocytes deprived of Notch signaling before β selection cannot progress to the DP stage (4, 5, 9, 11, 12). DN4 thymocytes also benefit from Notch ligand engagement en route to becoming DP cells (11). The importance of Notch activation at these stages has been demonstrated in vitro by culturing precursors either in the absence of Notch ligands or in the presence of pharmacological PS inhibitors, as well as in vivo by conditional gene deletion of CSL or Notch1, or induction of dnMAML. We now show that this is true also when thymocytes are genetically deprived of PS. Why Notch signaling is necessary in DN3/4 thymocytes is an open question. The difficulty lies in the paucity of data on direct target genes regulated by Notch in thymocytes and in the complex manner in which Notch mediates its effects (58). Notch signaling is an intricate, time-consuming, and context-dependent process that is initiated by ligand engagement and PS-mediated Notch cleavage at the cell surface, followed by transcriptional activation in the nucleus, resulting in changes in gene expression. Often, the genes regulated by Notch are themselves transcription factors, which in turn regulate the expression of other proteins. As it takes ≤1 d for β-selected DN3 thymocytes to differentiate to the DP stage (59), it is doubtful that the full effects of a Notch signal initiated in a late DN3/DN4 thymocyte happen within the same subset in which Notch ligand engagement and PS-mediated cleavage occurs. Thus, it is likely that Notch–Notch ligand interactions in late DN3/DN4 thymocytes influence the expression of many genes in late DN/early DP cells, including some that are important for TCR signaling and positive selection.
Direct target genes of Notch signaling are largely unknown, and are highly cell lineage– and stage-specific. To examine the effects of PS deficiency on a broad range of genes, we performed microarray analyses of RNA isolated from DP thymocytes (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070550/DC1). Despite the dramatic phenotype of PS1/2KO thymocytes, few genes were differentially expressed greater than twofold. This was consistent with data showing that blocking Notch hyperactivity in T-ALL cells results in no more than threefold differences in gene expression (60). In agreement with the flow cytometry data, PS1/2KO thymocytes showed reduced levels of CD5 mRNA, whereas mRNA for CD3, CD4, CD8, CD44, and CD69 were equivalent. No differences were observed in mRNA level for proteins involved in proximal TCR signaling, e.g., Lck and ZAP70. Consistent with a failure to detect increased apoptosis, the expression of antiapoptotic proteins Bcl-2 and -XL were equivalent. Because Notch can regulate the expression of transcription factors, e.g., c-Myc and c-Fos (60, 61), one intriguing difference in PS1/2KO thymocytes is the increased expression of AP-1 family members, a change that compliments the observation that Notch activity causes down-regulation of cFos (62). Altered expression of AP-1 proteins is notable because they influence cell cycle progression and differentiation, and extracellular signal-regulated kinase and AP-1 often regulate one another (63, 64). Moreover, in mature CD4 T cells, JunB is important in Th2 lineage cells, and Th1/2 responses can be altered in the absence of Notch (7, 20, 28, 65).
Although the most likely explanation for impaired TCR signaling in PS1/2KO thymocytes is altered gene expression, the possibility that Notch physically participates in TCR signaling cannot be excluded. Notch proteins are expressed by DP thymocytes (32, 66–69) and can be recruited to the site of TCR–MHC contact between DP thymocytes and APCs (32). Notch can bind PI3 kinase, Lck, and members of the ubiquitously expressed Deltex family (49, 70, 71). Because Deltex proteins are detected in the cytoplasm and have domains that can interact with both Notch and SH3 domain–containing proteins like Grb2, they have the potential to mediate clustering of these proteins at the membrane where proximal TCR signaling occurs. Notch has also been shown to physically associate with NF-κb and the IKK complex, and to influence NF-κb activity (30, 72).
Conditional PS1/2KO mice are the first Notch loss-of-function model reported to have defects in TCR signaling and MHC selection. Because thymocytes express all four Notch homologues, and the NICDs of Notch1–3 are highly conserved, it is not surprising that loss of any single Notch family member has no effect on thymocyte selection (16–19). It might be expected, however, that mutation of PS, dnMAML, and CSL would have similar effects. Nevertheless, no abnormalities in DP cellularity or in the development of SP thymocytes were reported with Cd4Cre-mediated RBPJκ gene deletion or dnMAML induction (7–20). The more severe effects of PS deficiency could reflect differences in the model systems, such as the half-life of PS, dnMAML, and CSL proteins. Both NICD and MAML are short-lived, but it is unclear whether CSL is degraded after Notch activation or if CSL remains permanently bound to DNA (58). Another possibility is differences in the timing of gene deletion. Cd4Cre-mediated deletion of the PS1 gene is detectable in a subset of DN2/DN3 thymocytes, and virtually no floxed PS1 remains by the DP stage. In contrast, it is not until thymocytes reach the DP stage that detectable levels of dnMAML-GFP are expressed in mice with Cd4Cre-mediated induction of dnMAML (20), and it is not until the DP stage that most thymocytes have undergone deletion of the RBPJκ gene by Cd4Cre in the conditional KO mice (Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20070550/DC1) (7). Although not discussed by the authors, mice with earlier deletion of CSL or expression of dnMAML appear to have alterations in SP thymocyte development (7, 12). Because both the CSL- and PS-deficient mice examined here were on mixed genetic backgrounds, it is unclear whether differences in the timing of gene deletion reflect differences in genetics and/or accessibility of the targeted loci. Taken with the fact that thymocytes deprived of Notch signaling before or during β-selection cannot mature past the DN3 stage, these data illustrate that the precise timing of Notch inhibition is a critical factor for revealing adverse effects on SP thymocyte development.
Although delayed loss of the RBPJκ gene may be the most likely explanation for why PS1/2KO mice have more severe phenotype than RBPJκ KO mice, it is notable that the disparities in the phenotype of CSL and PS mutant mice are reminiscent of findings from Drosophila, where CSL mutations can produce less severe phenotypes than those of Notch because Notch function is not restricted to relief of CSL-mediated transcriptional repression (73). Indeed, there are many examples of CSL-independent Notch functions. Truncated forms of Notch that cannot bind CSL retain activity (74–76). NICD can bind to and enhance the activity of transcription factors other than CSL (72, 74, 76, 77). Notch can indirectly modulate the transcription of CSL-independent genes via competition for transcriptional coactivators (78), a function that remains in tact in RBPJκ KO cells or dnMAML-expressing cells. Additional CSL-independent roles proposed for Notch include functions based on its interactions with proteins such as Deltex, NF-κB, and the IKK complex (70, 72, 79).
As well as promoting transcription of Notch target genes, additional functions could explain the phenotypic differences of PS, dnMAML, and CSL mutant mice and γ-secretase inhibitor–treated cultures. Loss of CSL may affect gene silencing (26) because it is a default repressor, MAML acts as a coactivator for several transcription factors (80, 81), and PS can regulate proteins other than Notch. The fact that PSs have multiple substrates has been a relatively minor concern because, with the exception of Notch, thymocytes do not express most of the known γ-secretase substrates, and the PS substrates present in thymocytes show no functional importance by gene deletion. Moreover, many papers describe the use of pharmacological γ-secretase inhibitors to prevent Notch signaling during thymocyte development, and to date, no Notch-independent effects have been reported. To the contrary, several lines of evidence support the contention that inhibition of Notch signaling is responsible for the phenotypes of PS1/2KO thymocytes reported here. Importantly, both the DN to DP block and TCR signaling defects can be corrected by introduction of NICD. Reciprocal to PS deficiency, wild-type DP thymocytes expressing a NICD Tg show enhanced TCR signal transduction (24, 33, 51). The phenotype of Tg NICD DP thymocytes suggests that positive selection and TCR signaling are improved in vivo. In vitro NICD Tg DP cells respond as well or better than wild-type thymocytes to TCR stimulation.
Although reciprocal results are observed for PS1/2KO and NICD Tg mice in respect to signaling in DP thymocytes, these mutations do not yield reciprocal results in respect to CD4/CD8 SP thymocyte development. Paradoxically, both show reduced development of CD4 SP cells. However, positive selection and SP generation is a complex, multistep process whereby the net TCR signal achieved by a DP thymocyte undergoing selection is determined by many factors, including TCR/MHC affinity, thymocyte–stromal cell adhesion, and thymocyte migration. Because Notch is a master regulator of gene expression, constitutive overexpression of NICD likely induces many changes not mediated by endogenous Notch.
At this time, we have no evidence that inhibiting endogenous Notch signaling by eliminating PS has any impact on CD4/CD8 lineage commitment. Aside from attenuating TCR signaling in DP and reducing SP thymocyte generation, CD4/CD8 lineage specification appears normal in conditional PS1/2KO mice. In addition to the TCR Tg data shown, selection of the diverse TCR repertoire shows normal MHC class I and class II dependence for CD8 and CD4 SP development, respectively (unpublished data). Although Notch is widely acknowledged for a direct role in binary cell fate decisions, our data suggest that PS effects on CD4/CD8 T cell fate are mediated through the ability of Notch activity to modulate TCR signaling. We propose that the quantity of TCR signaling is the primary CD4/CD8 cell fate determinant, whereas PS-dependent activation of Notch serves to modulate TCR signals and, in doing so, affects the outcome of positive selection. These findings are reminiscent of those from vulva development in Caenorhabditis elegans, where Notch reinforces a fate decision initially biased by the quantity of the epidermal growth factor–mitogen-activated kinase signal by creating a negative-feedback loop that accentuates small differences in signaling (82). The Notch and Ras signaling pathways also collaborate in Drosophila photoreceptor R7 specification (83), where Notch signals create a positive-feedback loop, leading to a stronger, more sustained Ras signal. In these examples, Notch reinforces a signaling pathway that is actually the primary determinant of cell fate. In an analogous way, PS-dependent Notch signaling may reinforce TCR signaling in thymocytes to influence positive selection, and in peripheral T cells, TCR and Notch signals may cooperate to determine Th1/2 lineage choice.
Collectively, our studies provide in vivo evidence that TCR and Notch signaling pathways are functionally linked, and support a model in which PS-dependent Notch activation influences positive selection and CD4/CD8 T cell development by modifying TCR signal strength. Precisely how the Notch and TCR pathways work together remains to be elucidated. Notch signaling could regulate transcription of components of TCR signal transduction and/or directly participate in proximal TCR signaling. Similar gaps exist regarding the mechanism by which Notch functions at other stages of lymphocyte development. For example, even though it is known that Notch signaling is required for commitment to the T cell lineage, transition from the DN to DP stage, and generation of marginal zone B cells, we still lack a detailed understanding of what genes are induced/repressed, what cell types receive the Notch signal, and in what cell types/tissues fate decisions are made. It is also largely unknown what factors regulate the pattern of gene expression that underlies selection at the DP stage. Even in TCRαβ transgenic mice where all DP thymocytes express the same TCR, only a subset become SP, indicating that factors other than TCR/MHC affinity play a critical role in determining which DP cells are positively selected. Given our observations, the expression patterns of PS, Notch1–4, and Notch ligands in the thymus, and the well-established role of Notch in influencing cell fate decisions; indeed, Notch could be regulating the genes that ultimately determine DP thymocyte selection. The future challenge will be to further dissect the complex interactions between Notch and other signaling pathways that allow Notch to exert lineage- and stage-specific effects across a broad range of cell types.
MATERIALS AND METHODS
Mice.
Floxed (f/f) PS1 (37) mutant and PS2-null mutant (35) mice have been previously described and were provided by R. Feng and J. Tsien (Princeton University, Princeton, NJ) and A. Bernstein and D. Donoviel (Mount Sinai Hospital, Toronto, Canada), respectively. PS1-null mutant mice were provided by S. Sisodia and P. Wong (Johns Hopkins University, Baltimore, MD) (84). pLck-Notch1IC9 transgenic mice were provided by E. Robey (University of California, Berkeley, Berkeley, CA) (14). huCd2-Cd5 transgenic mice were created and provided by P. Love (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD) (43). Cd5-null mutant mice (41) were created by Klaus Rajewsky and provided by Paul Love. Floxed RBPJκ mutant mice were produced by T. Honjo (85) and provided by L. Hennighausen (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD). H-2b C57BL/10 (B10) Rag2 and B6.SJL-Cd45a Rag1-null mutants, B6.SJL-Cd45a (also called Ptprca or Ly5a), transgenic B6 Cd4Cre and B10 or B10.A (H-2k) AND TCR, 5CC7 TCR, and F5 TCR mice were obtained from a National Institute of Allergy and Infectious Disease (NIAID) breeding contract with Taconic. All TCR transgenic mice used for this study were RAG deficient, so no endogenous TCRs were expressed. MHC-deficient (B6 Abβ/β2M-null mutant) mice were purchased from Taconic. Mice were bred and maintained under specific pathogen-free conditions in the NIAID Research Animal Facilities in accordance with American Association of Laboratory Animal Care specifications, and on protocols approved by the NIAID Animal Care and Use Committee.
Hematopoietic chimeras.
BM suspensions were depleted of mature T cells with antibodies to Thy1.2 (J1J) and CD5 (C3PO), plus low-toxicity rabbit complement (Cedarlane Laboratories). Mixed hematopoietic chimeras were made by reconstitution of sublethally irradiated RAG2-deficient recipients (500 rad, cesium source). H-2b PS2 KO (Cd45 b; the control for the PS1/2KO) and B6.SJL (Cd45a) BM cells were mixed at a ratio of 1:1; H-2b PS1/2KO (Cd45b) and B6.SJL (Cd45a) BM cells were mixed at a ratio of 10:1. To study thymocyte development in the absence of selection, lethally irradiated MHC-deficient recipients (1,000 rad, cesium source) were reconstituted with T cell–depleted BM cells. Irradiated mice were maintained on antibiotic water until analysis at 4–6 wk after reconstitution.
Lymphocyte isolation.
Thymocyte single-cell suspensions were generated using 100 μm nylon mesh (PGC Scientifics).
PCR.
Genotyping for the floxed PS1 allele was performed using primers and PCR conditions previously described (37). The deleted PS1 allele was detected with an additional primer, 5′-GCATATGAATACCATGTAGCTG-3′. To detect the 533-bp PCR product generated by the deleted PS1 allele, samples were amplified for an additional 10 cycles, for a total of 45 cycles. The floxed and deleted RBPJk alleles were detected using the following three primers: 5′-GATAGACCTTGGTTTGTTGTTTGG-3′, 5′-CCACTGTTGTGAACTGGCGTGG-3′, and 5′-AACATCCACAGCAGGCAAC-3′. PCR amplification of RBPJk floxed allele yields a PCR product of ∼500 bp, and the deleted RBPJk allele yields a product of ∼700 bp. Lineage-negative thymocytes were enriched by magnetic bead depletion using a lineage cocktail containing phycoerythrin (PE)-conjugated antibodies recognizing CD3ε, CD4, CD8α, CD11b, CD11c, CD19, CD45R, CD49b, CD122, Ter119, I-A, I-E, F4/80, Ly6G, TCRαβ, TCRγδ, and NK1.1 plus anti-PE beads and an AutoMACS (Miltenyi Biotec). Individual subsets of Lin− thymocytes were obtained by cell sorting for Lin−CD45+CD44+CD25+ (DN2), Lin−CD45+CD44−CD25+ (DN3), and Lin−CD45+CD44−CD25− (DN4) subsets. DP and SP thymocytes were isolated by cell sorting CD4+CD8+ (DP), TCRβhiCD4+ (CD4 SP), and TCRβhiCD8+ (CD8 SP).
RT-PCR.
RNA was isolated using Trizol (Invitrogen) according to the manufacturer's instructions. cDNA was generated using a SuperScript III First-strand Synthesis System for RT-PCR kit (Invitrogen) according to manufacturer's instructions. The primers used were Hes1 forward 5′-GCCAGTGTCAACACGACACCG-3′, Hes1 reverse 5′-TCACCTCGTTCATGCACTCG-3′, β-actin forward 5′-GTGGGCCGCTCTAGGCACCAA-3′, and β-actin reverse 5′- CTCTTTGATGTCACGCACGATTTC-3′.
Protein lysates and Western blotting.
Single-cell suspensions were lysed in buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 mM Na3VO4, and protease inhibitor tablets (Roche). Samples were incubated on ice for >30 min, inverted every 10 min to mix, and spun for 5 min at 12,000 rpm at 4°C. Supernatants were stored at −80°C.
Lysates were subjected to PAGE (Novex 12% NuPAGE gels) with MES buffer (Invitrogen), and transferred to Hybond-P polyvinylidene difluoride membranes (GE Healthcare). After blocking with 5% nonfat dried milk in PBS/0.01% Tween 20, blots were hybridized overnight at 4°C with antisera in PBS/0.01% Tween20 with 2.5% nonfat dried milk. Antibodies used were anti-PS1 (303–316; Oncogene Research Products/Calbiochem), anti–β-tubulin (H-235; Santa Cruz Biotechnology) and secondary anti–mouse-IgG-POD/anti–rabbit IgG-POD (Roche). Blots were developed with ECL Western Blotting Analysis System (GE Healthcare) or SuperSignal West Dura Extended Duration Substrate (Pierce Chemical Co.).
In vivo BrdU labeling.
Each mouse received a single i.p. injection with 1.8 mg BrdU (BD Biosciences) and was then maintained on drinking water supplemented with 0.8 mg/ml BrdU (Sigma-Aldrich) for up to 7 d. Data points for BrdU+ CD4+CD8+ DP or TCRhi CD4 and CD8 SP thymocytes are shown as a percentage of total thymocytes plotted against labeling time. Slopes of the lines were calculated using the linear curve function with a spreadsheet program (Excel; Microsoft). For DP thymocytes, only data points in the linear part of the curve (day 0–3) were used to calculate slopes.
Monoclonal antibodies.
The following monoclonal antibodies were purchesed from BD Biosience: anti-CD3ε-PE, -CyC, and -APC (145-2C11); anti–CD5-FITC and -PE(53–7.3); anti–CD4-CyC (RM4-5); anti–CD8α-CyC (53–6.7); anti–CD11c-PE (HL3); anti–CD19-PE (1D3); anti–CD25-FITC (7D4); anti–CD44-Biotin (IM7); anti–CD45-FITC (30-F11); anti–CD45.1-PE (A20); anti–CD45R-CyC (RA3-6B2); anti–CD49b-PE(DX5); anti–CD69-FITC (H1.2F3); anti–TCRγδ PE (GL3); anti–TCRαβ-FITC, -APC (H57.597), and -TCRVα11 (RR8-1); anti–NK1.1-PE (PK136); anti–Ter119-PE (TER-119); anti–I-Ab-PE(AF6-120.1); anti–I-A/I-E-PE (M5/114.15.2); anti–BrdU-FITC, and -APC (3D4). The following were obtained from Caltag Laboratories: anti–CD8α-FITC, -PE (CT-CD8), and anti–CD45R-PE (RA3-6B2). The following were purchased from eBioscience: anti-CD11b-PE (M1/70); anti-CD44-APC (IM7); anti-CD45.2-FITC, -PE, -PE-Cy5.5, and -APC-Cy5.5 (104); anti-CD45.1-FITC, -PE-Cy5.5, and -APC (A20); anti-CD122-PE (5H4); anti-F4/80-PE (BM8); anti-TCRαβ-PE (H57.597); anti-Ly6G (GR1)-PE (RB6-8C5). The following was obtained from Becton Dickinson Collaborative Technologies: anti-CD4-PE (GK1.5). Biotin-conjugated antibodies were visualized with Streptavidin-PE and -Cy5 (Jackson ImmunoResearch Laboratories).
Immunofluorescence analysis.
A single-cell suspension of lymphocytes in HBSS containing 0.2% BSA and 0.1% NaN3 (HBSS/BSA/NaN3) was incubated with properly diluted mAb at 4°C for 20 min. After staining, cells were washed twice with PBS/BSA/NaN3, and relative fluorescence intensities were measured by four-color fluorescence flow cytometry using a FACSCalibur flow cytometer with CellQuest software (both from Becton Dickinson), and FlowJo software (Tree Star, Inc.) was used for analysis. Staining for incorporated BrdU was performed using a BrdU Flow kit (BD Biosciences) according to the manufacturer's instructions.
Ca2+ measurements.
Thymocytes were washed and resuspended in HBSS with calcium and magnesium, 10 mM Hepes, and 1% FBS and probenecid. The calcium probe Indo-1 and detergent the Pluronic (Invitrogen) were added to cells and incubated for 30 min at 37°C. After washing, the cells were stained at room temperature with anti-CD8, -CD4, and -TCR. Cells were analyzed on an LSR II flow cytometer (Becton Dickinson) equipped with an argon laser tuned to 488 nm, a HeNe laser tuned to 633 nm, and a He-Cd UV laser tuned to 325 nm. Indo-1 fluorescence was analyzed using a 450-DC-LP dichroic beam splitter and emission filters at 395/20 and 530/30 nm for bound and free probes, respectively. The signals for bound and unbound Indo-1 were collected in linear mode. For each stimulation, an aliquot of cells in HBSS with 2 mM Ca (Biosource) was warmed at 37°C for 5 min before stimulation with azide-free, low-endotoxin, biotinylated anti-CD3 (2C11). Data were collected for 30 s, at which time 10 μg/ml Streptavidin (Sigma-Aldrich) was added. In some cases, 8 μg/ml ionomycin (Calbiochem) was added. To measure internal Ca2+ stores, cells were suspended in Ca2+-free HBSS with 0.5 mM EGTA; 10 μg/ml thapsigargin (Sigma-Aldrich) was added at 30 s, and data were collected for an additional 9.5 min. FACSDiva (Becton Dickinson) software was used for data acquisition, and FlowJo software (Tree Star, Inc.) was used for kinetic analysis.
Statistical analyses.
All two-tailed Student's t tests and linear regression analyses were conducted using Excel. Statistical analysis of BrdU labeling data were analyzed using Prism version 4.03 (GraphPad) for Windows. Error bars represent the SEM, where n ≥ 9 individual mice.
Online supplemental material.
Fig. S1 shows PS2 expression by thymocytes. Fig. S2 shows the effect of PS deficiency on MHC class II–restricted 5CC7 TCR Tg thymocytes. Fig. S3 shows the effect of PS deficiency on MHC class I–restricted F5 TCR Tg thymocytes. Fig. S4 shows changes in gene expression in control versus PS1/2° DP thymocytes. Fig. S5 shows timing of CD4Cre-mediated gene deletion in RBPJκflox thymocytes. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070550/DC1.
Supplemental Material
Acknowledgments
We appreciate the expert technical assistance of Sharron Evans, Rebecca Amos, and David Stephany. We thank Ellen Robey, Wendy Shores, Paul Love, and Nevil Singh for helpful discussion and comments on the manuscript.
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
The authors have no conflicting financial interests.
Abbreviations used: CSL, CBF1/Su(H)/Lag-1; dn, dominant negative. DN, double negative; dnMAML, dn Mastermind-like; DP, NIAID, National Institute of Allergy and Infectious Disease; NICD, Notch intracellular domain; PE, phycoerythrin; PS, Presenilin; SP, single positive; Tg, transgene.
References
- 1.Robey, E., and B.J. Fowlkes. 1994. Selective events in T cell development. Annu. Rev. Immunol. 12:675–705. [DOI] [PubMed] [Google Scholar]
- 2.Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. [DOI] [PubMed] [Google Scholar]
- 3.Bosselut, R. 2004. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat. Rev. Immunol. 4:529–540. [DOI] [PubMed] [Google Scholar]
- 4.Ciofani, M., T.M. Schmitt, A. Ciofani, A.M. Michie, N. Cuburu, A. Aublin, J.L. Maryanski, and J.C. Zuniga-Pflucker. 2004. Obligatory role for cooperative signaling by pre-TCR and notch during thymocyte differentiation. J. Immunol. 172:5230–5239. [DOI] [PubMed] [Google Scholar]
- 5.Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJβ rearrangement and allows pre-TCR-independent survival of early αβ lineage thymocytes. Immunity. 16:869–879. [DOI] [PubMed] [Google Scholar]
- 6.Washburn, T., E. Schweighoffer, T. Gridley, D. Chang, B.J. Fowlkes, D. Cado, and E. Robey. 1997. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 88:833–843. [DOI] [PubMed] [Google Scholar]
- 7.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of αβ/γδT cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 20:611–622. [DOI] [PubMed] [Google Scholar]
- 8.Radtke, F., I. Ferrero, A. Wilson, R. Lees, M. Aguet, and H.R. MacDonald. 2000. Notch1 deficiency dissociates the intrathymic development of dendritic cells and T cells. J. Exp. Med. 191:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt, T.M., M. Ciofani, H.T. Petrie, and J.C. Zuniga-Pflucker. 2004. Maintenance of T cell specification and differentiation requires recurrent Notch receptor–ligand interactions. J. Exp. Med. 200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciofani, M., G.C. Knowles, D.L. Wiest, H. von Boehmer, and J.C. Zuniga-Pflucker. 2006. Stage-specific and differential notch dependency at the αβ and γδT lineage bifurcation. Immunity. 25:105–116. [DOI] [PubMed] [Google Scholar]
- 11.Garbe, A.I., A. Krueger, F. Gounari, J.C. Zuniga-Pflucker, and H. von Boehmer. 2006. Differential synergy of Notch and T cell receptor signaling determines αβ versus γδ lineage fate. J. Exp. Med. 203:1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maillard, I., L. Tu, A. Sambandam, Y. Yashiro-Ohtani, J. Millholland, K. Keeshan, O. Shestova, L. Xu, A. Bhandoola, and W.S. Pear. 2006. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre–T cell receptor. J. Exp. Med. 203:2239–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowlkes, B.J., and E.A. Robey. 2002. A reassessment of the effect of activated Notch1 on CD4 and CD8 T cell development. J. Immunol. 169:1817–1821. [DOI] [PubMed] [Google Scholar]
- 14.Robey, E., D. Chang, A. Itano, D. Cado, H. Alexander, D. Lans, G. Weinmaster, and P. Salmon. 1996. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 87:483–492. [DOI] [PubMed] [Google Scholar]
- 15.Witt, C.M., V. Hurez, C.S. Swindle, Y. Hamada, and C.A. Klug. 2003. Activated Notch2 potentiates CD8 lineage maturation and promotes the selective development of B1 B cells. Mol. Cell. Biol. 23:8637–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch I in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 17.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, and K. Kumano. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 18:675–685. [DOI] [PubMed] [Google Scholar]
- 18.Kitamoto, T., K. Takahashi, H. Takimoto, K. Tomizuka, M. Hayasaka, T. Tabira, and K. Hanaoka. 2005. Functional redundancy of the Notch gene family during mouse embryogenesis: analysis of Notch gene expression in Notch3-deficient mice. Biochem. Biophys. Res. Commun. 331:1154–1162. [DOI] [PubMed] [Google Scholar]
- 19.Krebs, L.T., Y. Xue, C.R. Norton, J.R. Shutter, M. Maguire, J.P. Sundberg, D. Gallahan, V. Closson, J. Kitajewski, R. Callahan, et al. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 20.Tu, L., T.C. Fang, D. Artis, O. Shestova, S.E. Pross, I. Maillard, and W.S. Pear. 2005. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 202:1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French, M.B., U. Koch, R.E. Shaye, M.A. McGill, S.E. Dho, C.J. Guidos, and C.J. McGlade. 2002. Transgenic expression of numb inhibits notch signaling in immature thymocytes but does not alter T cell fate specification. J. Immunol. 168:3173–3180. [DOI] [PubMed] [Google Scholar]
- 22.Hadland, B.K., N.R. Manley, D. Su, G.D. Longmore, C.L. Moore, M.S. Wolfe, E.H. Schroeter, and R. Kopan. 2001. γ-Secretase inhibitors repress thymocyte development. Proc. Natl. Acad. Sci. USA. 98:7487–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doerfler, P., M.S. Shearman, and R.M. Perlmutter. 2001. Presenilin-dependent γ-secretase activity modulates thymocyte development. Proc. Natl. Acad. Sci. USA. 98:9312–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deftos, M.L., E. Huang, E.W. Ojala, K.A. Forbush, and M.J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 13:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellavia, D., A.F. Campese, E. Alesse, A. Vacca, M.P. Felli, A. Balestri, A. Stoppacciaro, C. Tiveron, L. Tatangelo, M. Giovarelli, et al. 2000. Constitutive activation of NF-kappa B and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 19:3337–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray, S., and M. Furriols. 2001. Notch pathway: making sense of suppressor of Hairless. Curr. Biol. 11:R217–R221. [DOI] [PubMed] [Google Scholar]
- 27.Adler, S.H., E. Chiffoleau, L. Xu, N.M. Dalton, J.M. Burg, A.D. Wells, M.S. Wolfe, L.A. Turka, and W.S. Pear. 2003. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J. Immunol. 171:2896–2903. [DOI] [PubMed] [Google Scholar]
- 28.Amsen, D., J.M. Blander, G.R. Lee, K. Tanigaki, T. Honjo, and R.A. Flavell. 2004. Instruction of distinct CD4 T helper cell fates by different Notch ligands on antigen-presenting cells. Cell. 117:515–526. [DOI] [PubMed] [Google Scholar]
- 29.Palaga, T., L. Miele, T.E. Golde, and B.A. Osborne. 2003. TCR-mediated Notch signaling regulates proliferation and IFNγ production in peripheral T Cells. J. Immunol. 171:3019–3024. [DOI] [PubMed] [Google Scholar]
- 30.Shin, H.M., H.M. Minter, O.H. Cho, S. Gottipati, A.H. Fauq, T.E. Golde, G.E. Sonenshein, and B.A. Osborne. 2006. Notch1 augments NF-kB activity by facilitating its nuclear retention. EMBO J. 25:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eagar, T.N., Q. Tang, M. Wolfe, Y. He, W.S. Pear, and J.A. Bluestone. 2004. Notch 1 signaling regulates peripheral T cell activation. Immunity. 20:407–415. [DOI] [PubMed] [Google Scholar]
- 32.Anderson, A.C., E.A. Kitchens, S.W. Chan, C.S. Hill, Y.N. Jan, W.M. Zhong, and E.A. Robey. 2005. The Notch regulator Numb links the Notch and TCR signaling pathways. J. Immunol. 174:890–897. [DOI] [PubMed] [Google Scholar]
- 33.Huang, Y.H., D. Li, A. Winoto, and E.A. Robey. 2004. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc. Natl. Acad. Sci. USA. 101:4936–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izon, D.J., J.A. Punt, L. Xu, F.G. Karnell, D. Allman, P.S. Myung, N.J. Boerth, J.C. Pui, G.A. Koretzky, and W.S. Pear. 2001. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 14:253–264. [DOI] [PubMed] [Google Scholar]
- 35.Donoviel, D.B., A.K. Hadjantonakis, M. Ikeda, H. Zheng, P.S.G. Hyslop, and A. Bernstein. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herreman, A., D. Hartmann, W. Annaert, P. Saftig, K. Craessaerts, L. Serneels, L. Umans, V. Schrijvers, F. Checler, H. Vanderstichele, et al. 1999. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. USA. 96:11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng, R., C. Rampon, Y.P. Tang, D. Shrom, J. Jin, M. Kyin, B. Sopher, G.M. Martin, S.H. Kim, and R.B. Langdon. 2001. Deficient neurogenesis in forebrain-specific presenilin-1 knockout mice is associated with reduced clearance of hippocampal memory traces. Neuron. 32:911–926. [DOI] [PubMed] [Google Scholar]
- 38.Lee, P.P., D.R. Fitzpatrick, C. Beard, H.K. Jessup, S. Lehar, K.W. Makar, M. Perez-Melgosa, M.T. Sweetser, M.S. Schlissel, and S. Nguyen. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. [DOI] [PubMed] [Google Scholar]
- 39.Huesmann, M., B. Scott, P. Kisielow, and H. von Boehmer. 1991. Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 66:533–540. [DOI] [PubMed] [Google Scholar]
- 40.Itano, A., and E. Robey. 2000. Highly efficient selection of CD4 and CD8 lineage thymocytes supports an instructive model of lineage commitment. Immunity. 12:383–389. [DOI] [PubMed] [Google Scholar]
- 41.Tarakhovsky, A., S.B. Kanner, J. Hombach, J.A. Ledbetter, W. Muller, N. Killeen, and K. Rajewsky. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 269:535–537. [DOI] [PubMed] [Google Scholar]
- 42.Pena-Rossi, C., L.A. Zuckerman, J. Strong, J. Kwan, W. Ferris, S. Chan, A. Tarakhovsky, A.D. Beyers, and N. Killeen. 1999. Negative regulation of CD4 lineage development and responses by CD5. J. Immunol. 163:6494–6501. [PubMed] [Google Scholar]
- 43.Azzam, H.S., J.B. DeJarnette, K. Huang, R. Emmons, C.S. Park, C.L. Sommers, D. El-Khoury, E.W. Shores, and P.E. Love. 2001. Fine tuning of TCR signaling by CD5. J. Immunol. 166:5464–5472. [DOI] [PubMed] [Google Scholar]
- 44.Yelon, D., and L.J. Berg. 1997. Structurally similar TCRs differ in their efficiency of positive selection. J. Immunol. 158:5219–5228. [PubMed] [Google Scholar]
- 45.Azzam, H.S., A. Grinberg, K. Lui, H. Shen, E.W. Shores, and P.E. Love. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayden-Martinez, K., L.P. Kane, and S.M. Hedrick. 2000. Effects of a constitutively active form of calcineurin on T cell activation and thymic Selection. J. Immunol. 165:3713–3721. [DOI] [PubMed] [Google Scholar]
- 47.Mariathasan, S., M.F. Bachmann, D. Bouchard, T. Ohteki, and P.S. Ohashi. 1998. Degree of TCR internalization and Ca2+ flux correlates with thymocyte selection. J. Immunol. 161:6030–6037. [PubMed] [Google Scholar]
- 48.Deftos, M.L., Y.W. He, E.W. Ojala, and M.J. Bevan. 1998. Correlating notch signaling with thymocyte maturation. Immunity. 9:777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sade, H., S. Krishna, and A. Sarin. 2004. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J. Biol. Chem. 279:2937–2944. [DOI] [PubMed] [Google Scholar]
- 50.Puls, K.L., K.A. Hogquist, N. Reilly, and M.D. Wright. 2002. CD53, a thymocyte selection marker whose induction requires a lower affinity TCR-MHC interaction than CD69, but is up-regulated with slower kinetics. Int. Immunol. 14:249–258. [DOI] [PubMed] [Google Scholar]
- 51.Laky, K., C. Fleischacker, and B.J. Fowlkes. 2006. TCR and Notch signaling in CD4 and CD8 T cell development. Immunol. Rev. 209:274–283. [DOI] [PubMed] [Google Scholar]
- 52.Matechak, E.O., N. Killeen, S.M. Hedrick, and B.J. Fowlkes. 1996. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 4:337–347. [DOI] [PubMed] [Google Scholar]
- 53.Vasquez, N.J., J. Kaye, and S.M. Hedrick. 1992. In vivo and in vitro clonal deletion of double-positive thymocytes. J. Exp. Med. 175:1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao, X.C., D.R. Littman, and A. Weiss. 1997. Itk and Fyn make independent contributions to T cell activation. J. Exp. Med. 186:2069–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaeffer, E.M., C. Broussard, J. Debnath, S. Anderson, D.W. McVicar, and P.L. Schwartzberg. 2000. Tec family kinases modulate thresholds for thymocyte development and selection. J. Exp. Med. 192:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lucas, J.A., A.T. Miller, L.O. Atherly, and L.J. Berg. 2003. The role of Tec family kinases in T cell development and function. Immunol. Rev. 191:1119–1138. [DOI] [PubMed] [Google Scholar]
- 57.Fischer, A.M., C.D. Katayama, G. Pages, J. Pouyssegur, and S.M. Hedrick. 2005. The Role of Erk1 and Erk2 in Multiple Stages of T Cell Development. Immunity. 23:431–443. [DOI] [PubMed] [Google Scholar]
- 58.Bray, S.J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689. [DOI] [PubMed] [Google Scholar]
- 59.Vasseur, F., A. Le Campion, and C. Pénit. 2001. Scheduled kinetics of cell proliferation and phenotypic changes during immature thymocyte generation. Eur. J. Immunol. 31:3038–3047. [DOI] [PubMed] [Google Scholar]
- 60.Palomero, T., W.K. Lim, D.T. Odom, M.L. Sulis, P.J. Real, A. Margolin, K.C. Barnes, J. O'Neil, D. Neuberg, A.P. Weng, et al. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 103:18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng, A.P., J.M. Millholland, Y. Yashiro-Ohtani, M.L. Arcangeli, A. Lau, C. Wai, C. del Bianco, C.G. Rodriguez, H. Sai, J. Tobias, et al. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Talora, C., D.C. Sgroi, C.P. Crum, and G.P. Dotto. 2002. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 16:2252–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hess, J., P. Angel, and M. Schorpp-Kistner. 2004. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117:5965–5973. [DOI] [PubMed] [Google Scholar]
- 64.Chalmers, C.J., R. Gilley, H.N. March, K. Balmanno, and S.J. Cook. 2007. The duration of ERK1/2 activity determines the activation of c-Fos and Fra-1 and the composition and quantitative transcriptional output of AP-1. Cell. Signal. 19:695–704. [DOI] [PubMed] [Google Scholar]
- 65.Minter, L.M., D.M. Turley, P. Das, H.M. Shin, I. Joshi, R.G. Lawlor, O.H. Cho, T. Palaga, S. Gottipati, J.C. Telfer, et al. 2005. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat. Immunol. 6:680–688. [PubMed] [Google Scholar]
- 66.Anderson, G., J. Pongracz, S. Parnell, and E.J. Jenkinson. 2001. Notch ligand-bearing thymic epithelial cells initiate and sustain Notch signaling in thymocytes independently of T cell receptor signaling. Eur. J. Immunol. 31:3349–3354. [DOI] [PubMed] [Google Scholar]
- 67.Felli, M.P., M. Maroder, T.A. Mitsiadis, A.F. Campese, D. Bellavia, A. Vacca, R.S. Mann, L. Frati, U. Lendahl, A. Gulino, and I. Screpanti. 1999. Expression pattern of Notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand- receptor interactions in intrathymic T cell development. Int. Immunol. 11:1017–1025. [DOI] [PubMed] [Google Scholar]
- 68.Huang, E.Y., A.M. Gallegos, S.M. Richards, S.M. Lehar, and M.J. Bevan. 2003. Surface expression of Notch1 on thymocytes: correlation with the double-negative to double-positive transition. J. Immunol. 171:2296–2304. [DOI] [PubMed] [Google Scholar]
- 69.Visan, I., J.B. Tan, J.S. Yuan, J.A. Harper, U. Koch, and C.J. Guidos. 2006. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 7:634–643. [DOI] [PubMed] [Google Scholar]
- 70.Diederich, R., K. Matsuno, H. Hing, and S. Artavanis-Tsakonas. 1994. Cytosolic interaction between deltex and Notch ankyrin repeats implicates deltex in the Notch signaling pathway. Development. 120:473–481. [DOI] [PubMed] [Google Scholar]
- 71.Matsuno, K., M. Ito, K. Hori, F. Miyashita, S. Suzuki, N. Kishi, S. Artavanis-Tsakonas, and H. Okano. 2002. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development. 129:1049–1059. [DOI] [PubMed] [Google Scholar]
- 72.Guan, E., J. Wang, J. Laborda, M. Norcross, P.A. Baeuerle, and T. Hoffman. 1996. T cell leukemia–associated human Notch/translocation-associated Notch homologue has IκB-like activity and physically interacts with nuclear factor-κB proteins in T cells. J. Exp. Med. 183:2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arias, A.M., V. Zecchini, and K. Brennan. 2002. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 12:524–533. [DOI] [PubMed] [Google Scholar]
- 74.Ross, D.A., and T. Kadesch. 2001. The Notch intracellular domain can function as a coactivator for LEF-1. Mol. Cell. Biol. 21:7537–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shawber, C., D. Nofziger, J.J. Hsieh, C. Lindsell, O. Bogler, D. Hayward, and G. Weinmaster. 1996. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 122:3765–3773. [DOI] [PubMed] [Google Scholar]
- 76.Wilson-Rawls, J., J.D. Molkentin, B.L. Black, and E.N. Olson. 1999. Activated Notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol. Cell. Biol. 19:2853–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jehn, B.M., W. Bielke, W.S. Pear, and B.A. Osborne. 1999. Cutting edge: protective effects of Notch-1 on TCR-induced apoptosis. J. Immunol. 162:635–638. [PubMed] [Google Scholar]
- 78.Shiama, N. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230–236. [DOI] [PubMed] [Google Scholar]
- 79.Vilimas, T., J. Mascarenhas, T. Palomero, M. Mandal, S. Buonamici, F. Meng, B. Thompson, C. Spaulding, S. Macaroun, M.L. Alegre, et al. 2007. Targeting the NF-κB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 13:70–77. [DOI] [PubMed] [Google Scholar]
- 80.Zhao, Y., R.B. Katzman, L.M. Delmolino, I. Bhat, Y. Zhang, A. Germaniuk-Kurowska, H.V. Reddi, A. Solomon, M.S. Zeng, A. Kung, et al. 2007. The notch regulator maml1 interacts with p53 and functions as a coactivator. J. Biol. Chem. 282:11969–11981. [DOI] [PubMed] [Google Scholar]
- 81.Katada, T., M. Ito, Y. Kojima, S. Miyatani, and T. Kinoshita. 2006. XMam1, Xenopus Mastermind1, induces neural gene expression in a Notch-independent manner. Mech. Dev. 123:851–859. [DOI] [PubMed] [Google Scholar]
- 82.Yoo, A.S., and I. Greenwald. 2005. LIN-12/Notch Activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 310:1330–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sundaram, M.V. 2005. The love-hate relationship between Ras and Notch. Genes Dev. 19:1825–1839. [DOI] [PubMed] [Google Scholar]
- 84.Wong, P.C., H. Zheng, H. Chen, M.W. Becher, D.J. Sirinathsinghji, M. Trumbauer, H. Chen, D.L. Price, L.H. Vanderploeg, and S. Sisodia. 1997. Presenilin 1 is required for Notch1 DII1 expression in the paraxial mesoderm. Nature. 387:288–292. [DOI] [PubMed] [Google Scholar]
- 85.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.