Abstract
Systemic onset juvenile idiopathic arthritis (SoJIA) represents up to 20% of juvenile idiopathic arthritis. We recently reported that interleukin (IL) 1 is an important mediator of this disease and that IL-1 blockade induces clinical remission. However, lack of specificity of the initial systemic manifestations leads to delays in diagnosis and initiation of therapy. To develop a specific diagnostic test, we analyzed leukocyte gene expression profiles of 44 pediatric SoJIA patients, 94 pediatric patients with acute viral and bacterial infections, 38 pediatric patients with systemic lupus erythematosus (SLE), 6 patients with PAPA syndrome, and 39 healthy children. Statistical group comparison and class prediction identified genes differentially expressed in SoJIA patients compared with healthy children. These genes, however, were also changed in patients with acute infections and SLE. An analysis of significance across all diagnostic groups identified 88 SoJIA-specific genes, 12 of which accurately classified an independent set of SoJIA patients with systemic disease. Transcripts that changed significantly in patients undergoing IL-1 blockade were also identified. Thus, leukocyte transcriptional signatures can be used to distinguish SoJIA from other febrile illnesses and to assess response to therapy. Availability of early diagnostic markers may allow prompt initiation of therapy and prevention of disabilities.
Juvenile idiopathic arthritis (JIA) is an important cause of short- and long-term disability. The term JIA encompasses a heterogeneous group of diseases that is classified according to three major types of presentation: oligoarthritis, polyarthritis, and systemic onset JIA (SoJIA). Each of these groups has different prognosis and responds differently to available therapies (1–4), suggesting that their pathogenesis is also unique.
Children with SoJIA usually present with systemic symptoms, fever and/or rash, which precede the development of arthritis for weeks or even years. Once arthritis develops, these patients have a highly variable disease outcome. The overall prognosis correlates with the persistence of systemic symptoms and the number of joints involved 6 mo after the initial presentation (5–8). Because of lack of success with conventional treatment, up to 50% of patients with SoJIA continue to have active arthritis 5–10 yr after diagnosis (2, 9, 10). Because long-term disability is directly correlated with duration of active disease, this group has the most severe outcome and thus has represented the most serious challenge to pediatric rheumatologists.
We have recently shown that IL-1 is a major mediator of the inflammatory cascade underlying SoJIA (11). Indeed, IL-1Ra is an effective treatment for this disease (11–14). IL-1 is also involved in the pathogenesis of familial autoinflammatory syndromes (15–17), and blocking IL-1 with IL-1Ra resolves the clinical symptoms of patients carrying mutations in the NALP3/cryopyrin gene (familial cold urticaria, Muckle-Wells syndrome, and NOMID/CINCA) (18–21) and in the PSTPIP1 gene (PAPA syndrome, a familial autoinflammatory disease that causes pyogenic sterile arthritis, pyoderma gangrenosum, and acne) (22, 23).
The diagnosis of SoJIA is currently based on clinical findings and requires the presence of arthritis (24). Because this manifestation may take months to develop, one of the major remaining challenges is how to establish an early diagnosis. As the presenting symptoms (fever and/or rash) and laboratory tests (anemia, leukocytosis, thrombocytosis, and elevated erythrocyte sedimentation rate) are nonspecific, patients undergo extensive diagnostic tests and hospitalizations to exclude infections and malignancies. The availability of an effective treatment fosters the need for diagnostic markers that will permit the initiation of therapy at an early stage of the disease to minimize the risk of developing long-term disabilities.
We have previously shown that microarray analyses of blood leukocytes from children with autoimmune diseases can be used to assess pathogenesis (25, 26). Here, we describe the use of blood leukocyte gene expression patterns to help diagnose patients with SoJIA during the systemic phase of the disease and to follow their response to therapy.
RESULTS
Patient characteristics
We analyzed 23 samples from 16 SoJIA patients displaying systemic symptoms (fever and/or rash) and arthritis, and 3 SoJIA patients with only systemic symptoms (fever, rash, and/or pericarditis) at the time of blood draw. Four patients (Sys12, Sys21, Sys25, and Sys51) were analyzed twice during independent systemic flares separated by 7–23 mo. Eight samples from patients with long-standing disease were obtained at the time of systemic disease flare (fever onset <4 preceding weeks). Five patients were newly diagnosed and also had had disease symptoms for no longer than 4 wk. SoJIA patients were predominantly females (female/male, 15/4). There were 10 Caucasian, 7 Hispanic, 1 Asian, and 1 African-American children in this group. Seven patients were not receiving any medication other than nonsteroidal antiinflammatory drugs at the time of blood draw. The remaining patients were receiving treatment with oral prednisone and/or IV methylprednisolone pulses, methotrexate, and/or anti-TNF therapy (Table S1, available at http://www.jem.org/cgi/content/full/jem.20070070/DC1). However, none of the treated patients had received IV pulses (methylprednisolone or infliximab) for at least 4 wk before blood draw. All patients fulfilled the International League of Associations for Rheumatology clinical diagnostic criteria for SoJIA (24) at the time of blood draw, during previous disease flares, or after blood draw. The average time from initiation of symptoms to establishment of diagnosis and initiation of therapy in these patients was 4.8 mo (range, 6 wk–3 yr). If the patient with the longest time to diagnosis (3 yr) is excluded, the average time to diagnosis for the remaining patient group was 2.7 mo. Two additional febrile patients (Sys 85 and Sys 91; Table S1) were studied but later proved to have a different diagnosis. Samples from 11 patients with established diagnosis of SoJIA in whom the systemic symptoms had subsided but arthritis persisted (SoJIA 2) and 14 patients with no systemic symptoms and no arthritis at the time of blood draw (SoJIA 3) were also analyzed. The demographic and clinical characteristics of these patients are summarized in Table S1. 20 ml of blood was drawn after informed consent. PBMCs were isolated within 2–4 h and processed and analyzed using Affymetrix human U133A and U133B GeneChips.
Blood leukocyte signatures differentiate SoJIA patients from healthy children
To identify genes whose expression would differentiate SoJIA patients (n = 16) from healthy controls (n = 16), statistical group comparisons were performed using the nonparametric Mann-Whitney rank test (P < 0.01) and Bonferroni correction. Transcripts displaying statistically significant differences (n = 873, 398 up-regulated and 475 down-regulated) were ordered by hierarchical clustering (Fig. 1 and Table S2, which is available at http://www.jem.org/cgi/content/full/jem.20070070/DC1). The 50 most significant genes are listed in Table S2 (marked with an asterisk). The expression of some of these genes can be interpreted based on our current knowledge of the disease. Related to the frequent anemia and the presence of erythroblasts in the blood of these patients, many erythroid lineage-specific genes are found up-regulated. Likewise, neutrophil-specific genes and genes that promote neutrophil survival (i.e., Foxo3a) (27) are found overexpressed. This is consistent with the neutrophilia present in SoJIA patients. Many of the over- and underexpressed genes, however, cannot be linked to a particular cell type or function.
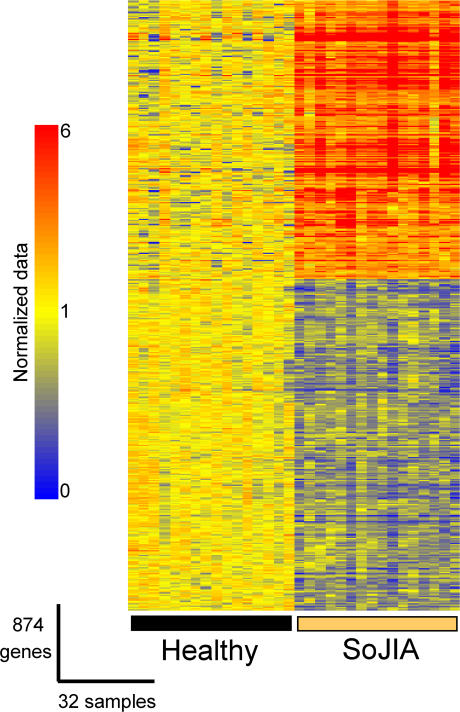
Figure 1.
Differential gene expression in PBMCs isolated from SoJIA patients and healthy controls. 17,454 genes passing the control criteria were tested. Genes expressed at statistically different levels between the two groups (P < 0.01; Wilcoxon-Mann-Whitney test, Bonferroni correction) were rearranged by hierarchical clustering to reveal differential expression. Expression values are normalized per gene to the healthy group. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression. A list of the genes shown in this figure is available in Table S2.
To identify a potential diagnostic signature, a two-step class prediction analysis was performed. It consisted of the identification of classifier genes and the validation of these genes in an independent cohort of patients and controls.
A subset of eight healthy volunteers and eight patients with an established diagnosis of SoJIA was used as a training set to identify the classifier genes (Fig. 2 A, left). The 50 most significantly differentially expressed transcripts (Table I) were then evaluated within the same set of patients in a leave-one-out cross-validation scheme. Using this strategy, 100% of the healthy and 88% of the SoJIA samples were classified accurately (seven were predicted accurately, and the class of the remaining sample was indeterminate). The ability of these transcripts to classify a test set composed of eight new healthy and eight independent SoJIA patients was tested. Using this approach, 100% of the patients and controls were accurately classified (Fig. 2 A, right).
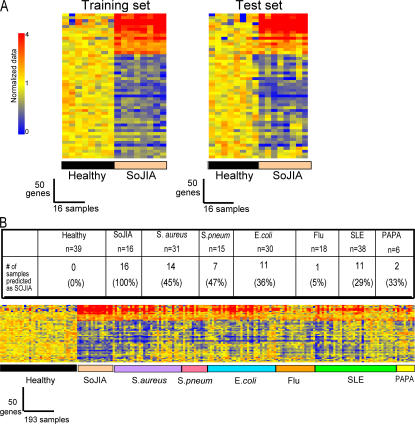
Figure 2.
Class prediction. (A) (Left) Eight healthy and eight SoJIA samples obtained from our initial study group were used as a training set to generate a list of classifier genes displaying the best ability to discriminate patients from healthy controls. In our training set, 100% of healthy and 88% of SoJIA patients were classified accurately. (Right) Those classifier genes were then tested on a test set (eight Healthy and eight SoJIA), and 100% of patients were classified accurately. Expression values were normalized per gene to the healthy group. Samples and genes were arranged by hierarchical clustering. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression. The list of the genes from this figure is shown in Table I. (B) Specificity of the SoJIA signature. The 50 best classifier genes from A were used to classify a test set of 39 healthy controls, 16 SoJIA, 31 S. aureus, 15 S. pneumoniae, 30 E. coli, 18 influenza A, 38 SLE patients, and 6 PAPA syndrome patients. The number of samples within each disease group predicted as SoJIA is represented on top of the figure. Genes were arranged by hierarchical clustering. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue relative low expression.
Table I.
50 classifiers distinguishing SoJIA patients from healthy controls
| Affymetrix ID | Gene symbol | p-value | Average normalized values in SoJIA |
Gene title |
|---|---|---|---|---|
| Protein biosynthesis | ||||
| 200002_at | RPL35 | 1.24E-08 | 0.6 | ribosomal protein L35 |
| 200089_s_at | RPL4 | 1.48E-10 | 0.6 | ribosomal protein L4 |
| 200802_at | SARS | 2.80E-09 | 0.8 | seryl-tRNA synthetase |
| 203113_s_at | EEF1D | 7.57E-11 | 0.2 | eukaryotic translation elongation factor 1 Δ (guanine nucleotide exchange protein) |
| 212018_s_at | RSL1D1 | 2.82E-10 | 0.5 | ribosomal L1 domain-containing 1 |
| 221726_at | RPL22 | 9.33E-10 | 0.7 | ribosomal protein L22 |
| Ubiquitination | ||||
| 209845_at | MKRN1 | 1.48E-10 | 4.2 | makorin, ring finger protein, 1 |
| 214790_at | SENP6 | 2.78E-09 | 0.5 | SUMO1/sentrin specific protease 6 |
| Microtubule/Cytoskeleton | ||||
| 210088_x_at | MYL4 | 7.57E-11 | 9.9 | myosin, light polypeptide 4, alkali; atrial, embryonic |
| 212878_s_at | KNS2 | 7.57E-11 | 0.6 | kinesin 2 60/70 kD |
| Transcription | ||||
| 200792_at | XRCC6 | 4.69E-09 | 0.7 | thyroid autoantigen 70 kD (Ku antigen) |
| 203617_x_at | ELK1 | 6.59E-07 | 1.6 | ELK1, member of ETS oncogene family |
| 204633_s_at | RPS6KA5 | 1.97E-08 | 0.6 | ribosomal protein S6 kinase, 90 kD, polypeptide 5 |
| 209430_at | BTAF1 | 1.57E-07 | 0.6 | BTAF1 RNA polymerase II, B-TFIID transcription factor–associated, 170 kD |
| 210504_at | KLF1 | 7.57E-11 | 6.7 | Kruppel-like factor 1 (erythroid) |
| 214177_s_at | PBXIP1 | 9.33E-10 | 0.6 | pre-B cell leukemia transcription factor interacting protein 1 |
| 217729_s_at | AES | 2.80E-09 | 0.5 | amino-terminal enhancer of split |
| 218490_s_at | ZNF302 | 4.68E-07 | 0.5 | zinc finger protein 302 |
| 224518_s_at | ZNF559 | 4.70E-08 | 0.5 | zinc finger protein 559 |
| 226327_at | ZNF507 | 0.000972 | 0.7 | zinc finger protein 507 |
| 225845_at | ZBTB44 | 1.97E-08 | 0.6 | BTB (POZ) domain-containing 15 |
| Metabolism | ||||
| 201050_at | PLD3 | 2.69E-05 | 1.4 | phospholipase D3 |
| 212174_at | AK2 | 4.66E-09 | 0.7 | adenylate kinase 2 |
| 226344_at | ZMAT1 | 4.27E-06 | 0.6 | zinc finger, matrin type 1 |
| 235802_at | PLD4 | 7.57E-11 | 0.4 | chromosome 14 open reading frame 175 |
| Immune response/Inflammatory response | ||||
| 211734_s_at | FCER1A | 2.82E-10 | 0.3 | Fc fragment of IgE, high affinity I, receptor for; α polypeptide |
| Transport | ||||
| 200063_s_at | NPM1 | 1.48E-10 | 0.5 | nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| 210854_x_at | SLC6A8 | 7.57E-11 | 9.2 | solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| 218978_s_at | SLC25A37 | 2.82E-10 | 7.5 | mitochondrial solute carrier protein |
| Heme | ||||
| 206834_at | HBD | 7.57E-11 | 36.4 | hemoglobin, Δ |
| 219672_at | ERAF | 7.57E-11 | 30 | erythroid associated factor |
| Apoptosis | ||||
| 223518_at | DFFA | 2.82E-10 | 1.9 | DNA fragmentation factor, 45 kD, α polypeptide |
| Unknown | ||||
| 201537_s_at | DUSP3 | 0.000157 | 1.8 | dual specificity phosphatase 3 (vaccinia virus phosphatase VH1- related) |
| 203818_s_at | SF3A3 | 5.20E-10 | 0.7 | splicing factor 3a, subunit 3, 60 kD |
| 209068_at | HNRPDL | 7.57E-11 | 0.5 | heterogeneous nuclear ribonucleoprotein D–like |
| 212830_at | EGFL5 | 4.69E-09 | 1.9 | EGF-like domain, multiple 5 |
| 213804_at | INPP5B | 1.06E-07 | 0.5 | inositol polyphosphate-5-phosphatase, 75 kD |
| 217807_s_at | GLTSCR2 | 7.47E-06 | 0.7 | glioma tumor suppressor candidate region gene 2 |
| 218877_s_at | TRMT11 | 7.12E-08 | 0.5 | chromosome 6 open reading frame 75 |
| 220755_s_at | C6orf48 | 1.48E-10 | 0.6 | chromosome 6 open reading frame 48 |
| 221932_s_at | C14orf87 | 7.57E-11 | 10.7 | chromosome 14 open reading frame 87 |
| 223011_s_at | OCIAD1 | 5.20E-10 | 0.6 | OCIA domain-containing 1 |
| 223656_s_at | C1orf91 | 4.70E-08 | 1.5 | hypothetical protein RP4-622L5 |
| 225159_s_at | — | 1.06E-07 | 0.7 | — |
| 225180_at | TTC14 | 1.57E-07 | 0.6 | tetratricopeptide repeat domain 14 |
| 226544_x_at | MUTED | 2.11E-05 | 0.8 | muted homolog (mouse) |
| 226680_at | ZNFN1A5 | 3.06E-08 | 0.6 | zinc finger protein, subfamily 1A, 5 |
| 228122_at | LOC285331 | 0.000128 | 0.7 | hypothetical protein LOC285331 |
| 235587_at | LOC202781 | 7.57E-11 | 0.5 | hypothetical protein LOC202781 |
| 241863_x_at | CCDC39 | 1.74E-06 | 0.5 | Coiled-coil domain-containing 39 |
List of 50 classifier genes distinguishing SoJIA patients from healthy controls. Genes are grouped based on functional ontology.
Some of the up-regulated transcripts that best classify SoJIA patients encode proteins involved in heme synthesis (delta hemoglobin and erythroid-associated factor) and erythrocyte-specific transcription factors (Kruppel-like factor 1), which could be related to the presence of nucleated erythroid precursors in SoJIA blood as described above.
Lack of specificity of the SoJIA signature
Children with SoJIA present with severe systemic symptoms (fever and rash) that usually precede the development of arthritis from weeks to years. Thus, the main differential diagnosis at presentation is an infectious disease. The 50 best SoJIA classifier transcripts described above were next tested for their ability to discriminate SoJIA patients from patients with infections (31 patients with Staphylococcus aureus, 15 patients with Streptococcus pneumoniae, 30 patients with Escherichia coli, and 18 patients with influenza A). As controls for noninfectious disease and steroid treatment, we included a group of 38 pediatric systemic lupus erythematosus (SLE) patients and 6 patients with PAPA syndrome, an IL-1–mediated autoinflammatory disease. These 50 genes were also dysregulated in patients with inflammatory conditions, as 45% of S. aureus, 47% of S. pneumoniae, 36% of E. coli, 5% of Influenza A, 29% of SLE, and 33% of PAPA syndrome patients were incorrectly classified as SoJIA (Fig. 2 B). Some of the genes included in the 50 best classifiers were mildly dysregulated in patients with established SoJIA and persistent arthritis, but not systemic symptoms (SoJIA 2) and in asymptomatic patients (SoJIA 3) (Fig. S1A, available at http://www.jem.org/cgi/content/full/jem.20070070/DC1). These patients, however, were correctly discriminated from the patients with systemic disease (Fig. S1 B).
Thus, the comparison of transcripts from SoJIA PBMCs and healthy controls is insufficient to yield SoJIA-specific signatures. Furthermore, the blood transcription patterns of SoJIA patients during the systemic phase of the disease are closer to those of patients with infections than to those of SoJIA patients in later (arthritic) stages of the disease.
Identification of a specific SoJIA signature
To identify a diagnostic SoJIA signature, the transcription profiles of SoJIA patients with systemic symptoms were directly compared with all the other infectious/inflammatory conditions. However, a large proportion of the predictor genes identified using this approach was found expressed similarly in SoJIA patients and healthy controls (unpublished data). Because it is difficult to control potentially confounding factors such as age or sex when comparing more than two groups of patients, we adopted a different strategy (see Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070070/DC1, and reference 28 for details). First, we performed statistical comparisons between each group of patients (16 SoJIA, 16 S. aureus, 10 S. pneumoniae, 10 E. coli, 10 influenza A, and 16 SLE) and their respective control groups composed of age- and gender-matched healthy donors. The p-values obtained from each comparison were then subjected to selection criteria that permitted the identification of genes significantly changed in SoJIA patients versus their control group, and not in any of the other disease versus control groups.
A nonstringent statistical group comparison (nonparametric Mann-Whitney rank test; P < 0.01) between 16 SoJIA and 10 healthy control samples yielded 4,311 differentially expressed transcripts (Fig. 3 A). 88 of these transcripts were expressed with a p-value of >0.5 in all other diseases compared with their corresponding healthy control groups (Fig. 3 B and Table II). More than 50% of these genes (47/88) encode proteins with unknown function. Among those encoding known proteins, several are involved in microtubule/cytoskeleton reorganization, ubiquitination, cellular transport, apoptosis, metabolism, transcription, protein biosynthesis, and posttranslational protein modification (Table II). The gene tree corresponding to these differentially expressed transcripts in individual patients and controls is displayed in Fig. 3 C. Only 1 of these 88 best classifiers (AK2) overlapped with the 50 genes that best discriminated SoJIA patients from healthy controls (Tables I and II), confirming the lack of specificity of our initial approach.
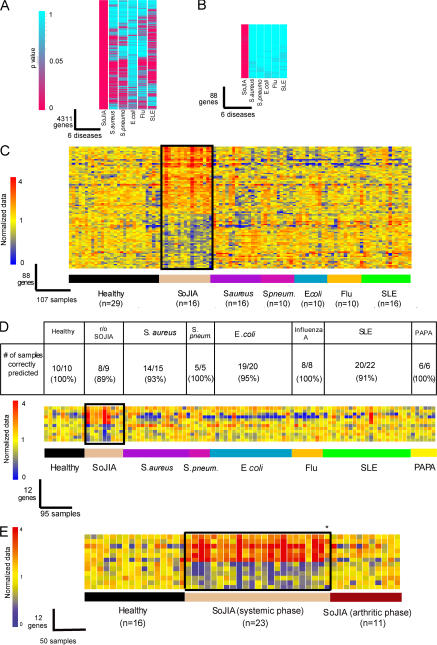
Figure 3.
SoJIA-specific signature. (A) Genes expressed at statistically different levels in the SoJIA patient group compared with healthy volunteers (P < 0.01; Wilcoxon-Mann-Whitney test) were selected (4,311 probe sets). p-values were similarly obtained from patients suffering from S. aureus, S. pneumoniae, E. coli, influenza A, and SLE. Each of these cohorts was compared with the corresponding control group. p-values are represented according to color scale: turquoise, low p-value; pink, high p-value. (B) 88/4,311 transcripts were found expressed at statistically different levels in the SoJIA patient group compared with healthy controls (P < 0.01; Wilcoxon-Mann-Whitney test), but not in all of the other groups compared with their healthy controls (P > 0.5; Wilcoxon-Mann-Whitney test). (C) The 88 genes from B were hierarchically clustered in the 107 samples used in the meta-analysis. Expression values of those 88 genes were normalized per gene to the healthy group. (D) The 12 most significant genes (P < 0.0001 in SoJIA group) were used as predictors for an independent test set of 10 healthy patients, 9 patients with febrile syndromes to rule out the diagnosis of SoJIA (2 of these patients were later on found to suffer from other diseases), 15 S. aureus, 5 S. pneumoniae, 20 E. coli, 8 influenza A, 22 SLE, and 6 PAPA syndrome patients. Expression values of those 12 genes were normalized per gene to the healthy group. Genes were arranged by hierarchical clustering. Transformed expression levels are indicated by color scale, with red representing relative high expression and blue indicating relative low expression. The list of the 88 and 12 genes shown in B–D is displayed in Table II. (E) The 12 gene signature displayed in D is shown in 16 healthy controls, 23 samples from 19 patients with established diagnosis of SoJIA while presenting systemic symptoms, and 11 patients with established diagnosis of SoJIA in whom the systemic symptoms had subsided but arthritis persisted. Only one patient with confirmed clinical diagnosis of SoJIA did not display the signature while having systemic symptoms (*).
Table II.
Best classifiers distinguishing SoJIA patients from infectious diseases, SLE, and PAPA
| Probe set ID | Gene symbol | p-value | Average normalized values in SoJIA |
Gene title |
|---|---|---|---|---|
| Apoptosis | ||||
| 212373_at | FEM1B | 5.27E-04 | 0.7 | Fem-1 homolog b (Caenorhabditis elegans) |
| 235116_at | TRAF1 | 9.06E-04 | 1.3 | TNF receptor–associated factor 1 |
| Extracellular matrix | ||||
| 202337_at | PMF1 | 9.06E-04 | 0.7 | polyamine-modulated factor 1 |
| 216993_s_at | COL11A2 | 0.00241 | 1.4 | collagen, type XI, α 2 |
| Glycosylation | ||||
| 201724_s_at | GALNT1 | 0.00462 | 0.9 | UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| 210205_at | B3GALT4 | 5.27E-04 | 1.3 | UDP-Gal:βGlcNAc β 1,3-galactosyltransferase, polypeptide 4 |
| Metabolism | ||||
| 209301_at | CA2 | 0.00374 | 2.6 | carbonic anhydrase II |
| 209509_s_at | DPAGT1 | 0.0015 | 1.2 | dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| 212174_at | AK2a | 8.80E-07 | 0.7 | adenylate kinase 2 |
| Microtubule/Cytoskeleton | ||||
| 200703_at | DNCL1 | 2.16E-04 | 1.7 | dynein, cytoplasmic, light polypeptide 1 |
| 207490_at | TUBA4 | 3.96E-04 | 1.4 | tubulin, α 4 |
| Nuclear mRNA splicing, via spliceosome | ||||
| 223416_at | SF3B14 | 0.00241 | 0.8 | splicing factor 3B, 14 kD subunit |
| 225394_s_at | MADP-1a | 2.62E-06 | 0.6 | MADP-1 protein |
| Phosphorylation | ||||
| 211992_at | WNK1 | 5.27E-04 | 2.1 | WNK lysine–deficient protein kinase 1 |
| 226979_at | MAP3K2 | 0.00567 | 0.7 | mitogen-activated protein kinase kinase kinase 2 |
| 227073_at | MAP3K2 | 0.00836 | 0.8 | mitogen-activated protein kinase kinase kinase 2 |
| Protein biosynthesis | ||||
| 212225_at | SUI1 | 2.16E-04 | 0.6 | putative translation initiation factor |
| 224302_s_at | MRPS36 | 0.00374 | 0.8 | mitochondrial ribosomal protein S36 |
| 226296_s_at | MRPS15a | 3.80E-05 | 0.6 | mitochondrial ribosomal protein S15 |
| Protein folding | ||||
| 201759_at | TBCD | 1.12E-04 | 2.2 | tubulin-specific chaperone d |
| 225061_at | DNAJA4 | 0.00191 | 2.4 | DnaJ (Hsp40) homolog, subfamily A, member 4 |
| 228622_s_at | DNAJC4a | 3.80E-05 | 0.7 | DnaJ (Hsp40) homolog, subfamily C, member 4 |
| Transcription | ||||
| 202484_s_at | MBD2 | 0.00191 | 0.7 | methyl-CpG–binding domain protein 2 |
| 224099_at | KCNH7 | 0.00191 | 1.5 | potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| 224933_s_at | JMJD1C | 0.00374 | 0.7 | jumonji domain-containing 1C |
| 225527_at | CEBPG | 0.00117 | 0.7 | CCAAT/enhancer-binding protein (C/EBP), γ |
| 227685_at | TMF1 | 0.0069 | 0.8 | TATA element modulatory factor 1 |
| 228785_at | ZNF281 | 0.00241 | 0.6 | Zinc finger protein 281 |
| 235389_at | PHF20 | 0.00462 | 0.8 | PHD finger protein 20 |
| 35671_at | GTF3C1 | 2.16E-04 | 1.3 | general transcription factor IIIC, polypeptide 1, α 220 kD |
| Transport | ||||
| 201066_at | CYC1 | 5.27E-04 | 0.8 | cytochrome c-1 |
| 202125_s_at | ALS2CR3 | 5.27E-04 | 2.1 | amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 3 |
| 213415_at | CLIC-2a | 1.69E-05 | 8.3 | CLIC-2 |
| 215716_s_at | ATP2B1 | 0.00241 | 0.6 | ATPase, Ca++ transporting, plasma membrane 1 |
| 218211_s_at | MLPH | 0.00462 | 1.5 | melanophilin |
| 224787_s_at | RAB18 | 6.94E-04 | 0.7 | RAB18, member RAS oncogene family |
| 225352_at | TLOC1a | 1.10E-05 | 2.4 | translocation protein 1 |
| 226154_at | DNM1L | 0.00836 | 0.8 | Dynamin 1–like |
| 238066_at | RBP7 | 0.00836 | 0.8 | retinol binding protein 7, cellular |
| 244227_at | SYT6 | 0.00241 | 1.3 | synaptotagmin VI |
| Ubiquitination | ||||
| 200718_s_at | SKP1A | 0.00462 | 1.3 | S-phase kinase–associated protein 1A (p19A) |
| 201824_at | RNF14 | 0.00301 | 2 | ring finger protein 14 |
| 210579_s_at | TRIM10 | 0.00835 | 1.4 | tripartite motif-containing 10 |
| Unknown | ||||
| 211994_at | WNK1a | 2.62E-06 | 2.8 | WNK lysine–deficient protein kinase 1 |
| 212055_at | C18orf10a | 5.54E-05 | 2 | chromosome 18 open reading frame 10 |
| 212341_at | MGC21416 | 0.00836 | 1.6 | hypothetical protein MGC21416 |
| 212829_at | — | 6.94E-04 | 2 | CDNA FLJ13267 fis, clone OVARC1000964 |
| 216739_at | — | 3.96E-04 | 1.6 | CDNA: FLJ20874 fis, clone ADKA02818 |
| 218116_at | C9orf78 | 0.00191 | 2.1 | chromosome 9 open reading frame 78 |
| 218126_at | FLJ10579 | 9.06E-04 | 1.5 | hypothetical protein FLJ10579 |
| 218583_s_at | RP42 | 0.00462 | 1.5 | RP42 homolog |
| 218936_s_at | HSPC128 | 0.00117 | 0.6 | HSPC128 protein |
| 222309_at | C6orf62 | 0.00567 | 0.6 | chromosome 6 open reading frame 62 |
| 223112_s_at | NDUFB10 | 3.96E-04 | 0.8 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 10, 22kDa |
| 223548_at | C1orf26 | 0.0015 | 1.4 | chromosome 1 open reading frame 26 |
| 224807_at | KIAA1533 | 0.0015 | 0.8 | KIAA1533 |
| 224915_x_at | TALDO1 | 9.06E-04 | 0.7 | transaldolase 1 |
| 225202_at | RHOBTB3 | 0.0069 | 1.2 | Rho-related BTB domain containing 3 |
| 225213_at | TA-PP2C | 2.16E-04 | 0.8 | T cell activation protein phosphatase 2C |
| 225819_at | TBRG1 | 0.00241 | 0.7 | transforming growth factor β regulator 1 |
| 226833_at | FLJ32499 | 0.00301 | 1.3 | hypothetical protein FLJ32499 |
| 226927_at | — | 0.00374 | 1.2 | hypothetical protein LOC728568 |
| 227265_at | FGL2 | 0.00301 | 0.8 | fibrinogen-like 2 |
| 228452_at | C17orf39 | 0.00625 | 1.6 | chromosome 17 open reading frame 39 |
| 228953_at | WHDC1a | 5.54E-05 | 0.6 | WAS protein homology region 2 domain containing 1 |
| 229074_at | EHD4 | 0.00117 | 0.8 | EH domain–containing 4 |
| 229653_at | FLJ10979 | 0.00836 | 1.4 | hypothetical protein FLJ10979 |
| 230118_at | — | 2.16E-04 | 1.3 | transcribed locus |
| 230421_at | LOC345462 | 0.00567 | 1.2 | similar to hypothetical protein 9630041N07 |
| 230546_at | VASH1a | 7.95E-05 | 1.6 | vasohibin 1 |
| 230747_s_at | C18orf17a | 3.80E-05 | 0.7 | chromosome 18 open reading frame 17 |
| 232486_at | LRFN1 | 0.00462 | 1.4 | leucine-rich repeat and fibronectin type III domain–containing 1 |
| 232709_at | — | 0.00191 | 0.7 | CDNA FLJ13427 fis, clone PLACE1002477 |
| 233469_at | psiTPTE22 | 0.00301 | 1.3 | TPTE pseudogene |
| 234305_s_at | MLZE | 9.06E-04 | 1.4 | melanoma-derived leucine zipper, extra-nuclear factor |
| 235798_at | — | 0.00117 | 0.8 | — |
| 236196_at | ZNF326 | 0.0015 | 0.7 | CDNA FLJ42548 fis, clone BRACE3004996 |
| 241491_at | KIAA1002 | 6.94E-04 | 1.5 | KIAA1002 protein |
| 241517_at | DDEF1 | 0.00117 | 1.3 | development and differentiation enhancing factor 1 |
| 241817_at | FLJ43654 | 3.96E-04 | 0.7 | FLJ43654 protein |
| 242003_at | LOC157697 | 0.00301 | 0.7 | hypothetical protein LOC157697 |
| 242300_at | UBB 3′ UTRa | 2.56E-05 | 4 | ubiquitin B (UBB) mRNA, 3′ UTR and genetic suppressor element |
| 243109_at | MCTP2 | 2.94E-04 | 1.7 | multiple C2-domains with two transmembrane regions 2 |
12 most significant genes used in the prediction.
To validate the microarray data, we selected the 8/88 best-characterized genes based on their genomic sequence to perform RT-PCR. RNA samples were obtained from 12 healthy controls (6 from our initial microarray analysis and 6 new ones) and 12 SoJIA, 5 S. aureus, 4 S. pneumoniae, 5 E. coli, and 5 influenza A patients (all from our initial microarray study). The results of these experiments confirmed that expression of these eight genes is significantly increased in SoJIA patients, but not in infections compared with healthy controls (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070070/DC1).
12 genes can be used to diagnose SoJIA
A more stringent analysis (P < 0.0001) permitted us to identify 12/88 genes highly differentially expressed in SoJIA compared with healthy controls, but not differentially expressed (P > 0.5) in all other disease groups compared with their respective controls. These 12 genes are included in Table II.
The ability of this set of 12 genes to identify patients with SoJIA was then validated using independent groups of patients and controls. A training set composed of 10 healthy and 16 SoJIA samples was used to predict sample class for an independent test set composed of a random group of 10 healthy patients, 9 patients with fever of more than 10 d in duration and negative bacterial cultures (suspected to have SoJIA), and 15 S. aureus, 5 S. pneumoniae, 20 E. coli, 8 influenza A, 22 SLE, and 6 PAPA syndrome patients (Fig. 3 D). This analysis allowed us to accurately classify six out of seven patients fulfilling SoJIA clinical diagnostic criteria. The only SoJIA patient not accurately classified (Sys 99 in Table S1) had fever, rash, and arthritis at the time of blood draw and eventually responded well to treatment with steroids and methotrexate. Two additional patients who were not classified as SoJIA (Sys 85 and Sys 91) were subsequently diagnosed with different diseases. Overall, 93% of S. aureus, 100% S. pneumoniae, 95% of E. coli, 91% of SLE, and 100% of influenza A and PAPA syndrome samples were correctly discriminated from SoJIA (Fig. 3 D). Thus, the “normalization” of each patient group to healthy control values and the comparison of significances rather than expression levels permitted us to extract a signature unique to the majority of SoJIA patients.
The expression of the 12 transcripts was next tested in the 11 SoJIA patients described above, in whom the systemic symptoms had subsided but continued to display arthritis. As shown in Fig. 3 E, 11/11 patients did not differentially express these genes compared with healthy controls, further indicating that this signature is specific to the systemic phase of the disease.
Whether the dysregulated expression of these 12 genes is related to the pathogenesis of SoJIA remains to be determined. Overall, seven of these genes encode uncharacterized proteins. Among those encoding proteins with known function, the most significantly up-regulated transcript (average, 8.2-fold), chloride intracellular channel 2 (CLIC-2), belongs to the ubiquitous glutathione transferase structural family. CLIC-2 is one of only a few cytosolic inhibitors of cardiac ryanodine receptor 2 channels and may suppress their activity during diastole and stress (29). Interestingly, CLIC-2 is the only transcript encoding a protein with potential link to IL-1 secretion, as chloride has been shown to play an important role in maintaining the P2X7 receptor (P2X7R) in a conformationally restrained state. This in turn limits the coupling of this receptor to signaling pathways that regulate caspase 1 and IL-1b signaling cascade (30).
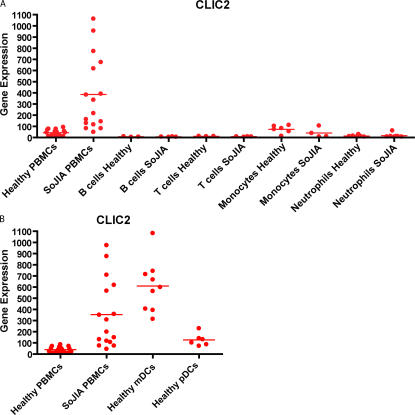
To determine if a specific cell type is preferentially contributing to the overrepresentation of these transcripts, their expression was analyzed in B cells, T cells, monocytes, and neutrophils from healthy donors and SoJIA patients, as well as myeloid DCs (mDCs) and plasmacytoid DCs (pDCs) from healthy donors. Interestingly, 5/12 transcripts showed the highest expression levels in mDCs (not depicted). Within the cells that we analyzed, expression of CLIC-2 is restricted to this cell type (Fig. 4, A and B). However, whether the 12 transcript signature derives from a cell type not normally present in the blood (i.e., a bone marrow precursor) needs to be ruled out.
Figure 4.
Transcription of CLIC-2 in blood cell subsets. (A) Total RNA was extracted from the PBMCs of 16 healthy donors and 16 SoJIA patients; B cells, T cells, and monocytes from 3 healthy donors and 4 SoJIA patients, and neutrophils from 6 healthy donors and 7 SoJIA patients. Amplified cRNA was hybridized on Affymetrix HG-U133 chips. Raw intensity values from each chip were first pre-scaled to the 500 target intensity value in Affymetrix Microarray suite before being imported and analyzed in GeneSpring 6.1. (B) Total RNA was extracted from the PBMCs of 16 healthy donors and 16 SoJIA patients, mDCs from 9 healthy donors, and pDCs from 6 healthy donors. Amplified cRNA was hybridized to Affymetrix HG-U133 chips. Raw intensity values from each chip were pre-scaled to the 500 target intensity value in Affymetrix Microarray suite before being imported and analyzed in GeneSpring 6.1.
Treatment with IL-Ra (Anakinra) partially extinguishes the SoJIA signature
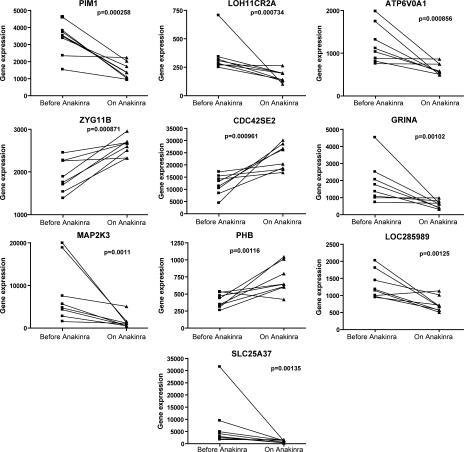
The effect of IL-1 blockade on the SoJIA-specific signature was analyzed by comparing the expression of the 873 transcripts that differentiate SoJIA patients from healthy controls (Fig. 1 and Table S2) in eight active SoJIA patients before and after initiation of IL-1 blockade. Anakinra was found to induce a significant change (P < 0.05) in the expression levels of 389 transcripts. The 10 most significant are displayed in Fig. 5. They include genes encoding proteins with antiapoptotic function (Pim-1) (31), lysosomal ATP-ase activity (ATP6V0A1), Cdc42-binding proteins linked to F-actin accumulation at the immunological synapse (Cdc42SE2) (32), glutamate receptor (GRINA), MAP2 kinase 3 (MAP2K3), prohibitin (PHB), and the solute carrier protein family (SLC25A37). Of them, only MAP2K3 has a known potential link to IL-1, as this molecule has been shown to down-modulate IL-1 responses on fibroblast-like synoviocytes and to regulate p38 activation in experimental inflammatory arthritis (33, 34).
Figure 5.
Anakinra effect on the specific SoJIA signature. Transcription levels (raw values) corresponding to the genes that best differentiate SoJIA patients from healthy controls (from Fig. 1 and Table S2) were analyzed before and after the initiation of Anakinra in eight patients. The 10 genes with most significant p-values are displayed (p-values were calculated using a paired, two-tailed t test).
Only 19 of the 88 diagnostic transcripts described above (Table II) overlapped with the 389 transcripts that significantly changed upon IL-1 blockade. Thus, the majority of transcripts that best differentiate SoJIA patients from patients with other inflammatory conditions do not appear to be the target of IL-1. This is not surprising, as IL-1 plays an important role in other diseases included in this analysis. Therefore, IL-1–related genes could have been counter-selected in our analysis of significance across diseases.
DISCUSSION
SoJIA is the only form of JIA in which systemic symptoms precede the appearance of joint inflammation. Because systemic symptoms and current laboratory tests are nonspecific, a major remaining challenge is how to establish the prompt diagnosis of the disease during the systemic phase to avoid lengthy hospitalizations and initiate effective therapy. Here, we show that gene expression patterns in blood leukocytes may be used, together with careful clinical assessment, to discriminate SoJIA during the systemic phase of the disease from other febrile conditions.
Patients with SoJIA display a very striking pattern of leukocyte gene transcription when compared with healthy controls. These differences might reflect changes in blood cell composition, i.e., the presence of a cell population not normally found in the blood, or fluctuations in numbers of blood-specific cell populations rather than real transcriptional changes. Active SoJIA patients, for example, display increased platelet and leukocyte numbers compared with healthy controls. Erythroid precursors, which are not normally present in peripheral blood, are also found in these patients, and erythroid-specific transcripts are among the most significantly up-regulated in SoJIA blood.
To control for changes in cell composition and find a SoJIA-specific signature, we included in our analysis 138 samples from pediatric patients with systemic inflammation, including bacterial and viral infections, autoimmune (SLE), and autoinflammatory (PAPA) diseases. Some of these patients (i.e., bacterial infections and PAPA syndrome) display alterations in blood cell numbers similar to those of SoJIA patients. As age, gender, and time of day when the blood is drawn have been described to influence blood gene expression patterns (35), our samples were matched for age and gender with controls and most of them were obtained within similar time frames.
There was a significant degree of overlap between the signatures of SoJIA patients and all the other inflammatory disease groups included in this study, especially Gram (+) bacterial infections, further stressing the need for multi-cohort studies when searching for disease-specific biomarkers. The overlap could be due to dysregulated cytokine production and/or signaling cascades that may be shared by some of these diseases. IL-1, for example, is important in the cascade of defense mechanisms against many bacterial infections (36, 37). Increased IL-1b production has also been described in autoinflammatory diseases including PAPA syndrome. Indeed, PAPA patients display a mutation in the PSTPIP1 gene (22) that exerts a dominant-negative effect on the activity of pyrin and leads to increased IL-1b production by peripheral blood leukocytes (16). Interestingly, the herein described blood transcription patterns of SoJIA patients during the systemic phase of the disease are closer to those of patients with infections than to those of SoJIA patients who have resolved the systemic phase but continue displaying active arthritis. This could be explained if different pathogenic events, i.e., different cytokine cascades, were responsible for the different phases of the disease. Alternatively, the same cytokine alteration in a more localized environment (i.e., the joint) may not give rise to a blood cell signature.
To identify a specific blood signature that would permit the accurate differential diagnosis of SoJIA patients during the systemic phase of the disease, we designed an analysis of significance across multiple febrile inflammatory diseases and control groups. One of the advantages of this analysis is that it permits us to normalize each disease group to its own matched control group, therefore avoiding biological (i.e., age, gender) or technical (i.e., array runs) confounding factors. Using this approach, a SoJIA-specific signature composed of 88 genes was identified. This signature is very stable over time, as we could identify it in two samples from a patient obtained >2 yr apart (unpublished data). Using a more stringent analysis (P < 0.0001 in SoJIA and P > 0.5 in all other groups), 12 highly significant genes permitted to accurately classify the disease in 18/19 SoJIA patients. Furthermore, it allowed us to rule out SoJIA in two febrile patients who later developed arthritis but proved to suffer from different diseases. It also allowed us to discriminate systemic infections with Gram (+) and Gram (−) bacteria in 48/50 patients. Perhaps more interesting, six out of six patients with PAPA syndrome were not classified as SoJIA. How this signature will perform in discriminating SoJIA patients from other autoinflammatory diseases where IL-1 is also dysregulated is currently being investigated.
Most of the genes included within the SoJIA-specific signature encode proteins that remain uncharacterized. Of those that encode known proteins, we did not find any obvious components of the IL-1 pathway. As discussed above, this might be expected as IL-1 also plays an important role in the inflammatory response against some of the infectious diseases included in this study. IL-1–related genes would have therefore been counter-selected. CLIC-2, the most up-regulated of the 12 transcripts that compose the SoJIA-specific signature, might be involved in the regulation of IL-1 secretion. This gene encodes a chloride channel, and chloride levels control the conformation of P2X7R (30). This in turn limits the coupling of this receptor to signaling pathways that regulate caspase 1 and IL-1b signaling cascade. Interestingly, expression of CLIC-2 within PBMCs seems to be restricted to mDCs (38). IL-1 amplifies DC function, and IL-1 production is induced when monocytes are co-cultured with alloreactive T cells and autologous DCs through a cell contact–dependent mechanism (39). Studies are currently underway to determine whether these genes are up-regulated in SoJIA blood mDCs or in a different cell population not normally present in the blood of healthy and infected children.
Even though the clinical symptoms were controlled in the majority of SoJIA patients treated with an IL-1 receptor antagonist, expression of only a fraction (389/873) of the dysregulated transcripts returned to control levels in treated patients. Several explanations might be put forward to explain this observation. IL-1 could be downstream of a factor present in SoJIA serum that is not blocked by Anakinra. Indeed, the residual SoJIA signature might be a tool to identify such an IL-1–inducing factor. The IL-1 antagonist effect of Anakinra may be sufficient to silence the clinical symptoms, but not to erase the molecular signature. Finally, our limited patient sample before and after Anakinra treatment may not be enough to give statistical power to this analysis, and more patients may need to be studied to draw firm conclusions.
The small number of genes identified in this study as SoJIA-specific might help in the diagnosis of patients with febrile conditions included under the term “fever of unknown origin.” They might also allow to promptly initiate specific therapy in SoJIA patients even before arthritis develops, thus avoiding the need for additional therapies and the development of long-term disabilities. The identification of genes whose expression is restored back to normal levels upon successful IL-1 blockade might also help identify predictors of response to therapy in longitudinal studies.
SoJIA patients are heterogeneous, however, regarding severity of symptoms and disease course (38, 40). Further studies will therefore be required to confirm the value of blood gene transcription profiling in establishing the diagnosis and predicting the response to IL-1 blockade in larger patient cohorts.
MATERIALS AND METHODS
Patient information.
Blood samples were obtained from 21 patients with clinical diagnosis of SoJIA (4 patients were tested twice at an interval of 1–2 yr) during the systemic phase of the disease (median age, 6 yr; range, 2–17 yr). In two patients, subsequent clinical course ruled out this diagnosis. Samples were from 11 patients with SoJIA in the arthritis phase of the disease, 14 asymptomatic patients, and 8 patients with SoJIA treated with Anakinra (6 yr, 2–19 yr). These were also analyzed together with 94 patients hospitalized with active infections: 30 patients with E. coli infection (1 yr; 2 wk–16 yr), 31 patients with S. aureus infection (7 yr; 3 mo–18 yr), 15 patients with S. pneumoniae (3.1 yr; 2 mo–16 yr), 18 with influenza A infections (1.5 yr; 3 wk–16 yr), 38 patients with SLE (12 yr; 5–18 yr), and 6 patients with PAPA syndrome (32.6 yr; 11–68 yr). Patients were divided in training and test sets according to age and treatment (Table S1). Subjects were recruited at Texas Scottish Rite Hospital and Children's Medical Center of Dallas. Healthy controls included patients with hypermobility attending the rheumatology clinic and healthy children undergoing routine surgical procedures with no history of asthma, acute infections, or inflammatory conditions. Most of the samples were obtained between 9:00 a.m. and 12:00 p.m. The study was approved by the Institutional Review Boards of UT Southwestern Medical Center, Texas Scottish Rite Hospital, and Baylor Health Care System, and informed consent was obtained from all patients (legal representatives and patients over 10 yr of age). Bacterial and viral infections were confirmed by standard bacterial cultures, direct fluorescent antigen testing, and viral cultures. Patients with infections were recruited once a confirmed microbiologic diagnosis was established. Respiratory viral cultures were performed in 60 of 73 (82%) patients with bacterial infections. The clinical and demographic characteristics of these patients are summarized in Table S1 and have been reported elsewhere (41). PAPA syndrome patients were diagnosed based on clinical data and sequencing the PSTPIP1 gene.
RNA and microarray sample preparation from PBMC and blood cell subpopulations.
All blood samples were obtained in EDTA purple-top tubes (BD Vaccutainer). Fresh PBMCs were isolated via Ficoll gradient. Cells were lysed in RLT lysis buffer containing β-mercaptoethanol (QIAGEN).
pDCs and mDCs were purified from a healthy donor's buffy coats. Ficoll-enriched PBMCs were depleted of lineage+ cells with CD3, CD14, CD19, CD16, CD56, and glycophorin A microbeads (Miltenyi Biotec). After staining with lineage cocktail-FITC, CD11c-allophycocyanin, and CD123-PE (BDBiosciences), and HLA-DR-QR (Sigma-Aldrich) mAbs, cells were sorted on a FACSVantage (Becton Dickinson) to at least 99% purity.
Monocytes, B cells, and T cells were positively selected from PBMCs using CD14 microbeads, CD19 microbeads, or CD3 microbeads (Miltenyi Biotec) and MS column (Miltenyi Biotec) according to the manufacturer's instructions to at least 95% purity. Neutrophils were isolated from venous blood of healthy volunteers by dextran (GE Healthcare) sedimentation of erythrocytes and density gradient centrifugation of leukocytes. The resulting cell populations contained <2% contaminating cells.
Total RNA was isolated using the RNeasy kit (QIAGEN) according to the manufacturer's instructions, and the RNA integrity was assessed by using an Agilent 2100 Bioanalyzer (Agilent). For PBMCs and neutrophils, 5 μg of total RNA was used to generate double-stranded cDNA containing the T7-dT (24) promoter sequence (Operon) as a template for in vitro transcription single-round amplification with biotin labels, using the Enzo R BioArrayTm HighYieldTM RNA Transcript Labeling kit (Affymetrix, Inc.). For the other cell subtypes, the same protocol was used starting with 50 ng of total RNA and performing two rounds of amplification. Biotinylated cRNA targets were purified using the Sample Cleanup Module (Affymetrix, Inc.) and hybridized to human U133A and B GeneChips (Affymetrix, Inc.) according to the manufacturer's standard protocols. Arrays were scanned using a laser confocal scanner (Agilent).
Microarray data analysis.
For each Affymetrix U133A and B GeneChip, raw intensity data were normalized to the mean intensity of all measurements on that array and scaled to a target intensity value of 500 (TGT) in Affymetrix Microarray Suite 5.0. Data were then further analyzed using GeneSpring software version 7.0. Data were normalized to a set of healthy controls (sex- and age-matched). An Affymetrix flag of “present” in at least 75% of samples of each cohort designated the filter of reliable intensity measurement from each individual gene chip. The combined two lists (17,231 probes) were used as quality control for statistical tests, class prediction, and clustering algorithms subsequently performed on the data. Class comparison was performed using nonparametric ranking statistical analysis test (Mann-Whitney) applied to quality control genes. In the vertical direction, hierarchical clusters of genes were generated using the Spearman correlation. Class prediction was performed using a supervised learning algorithm, K-Nearest Neighbors Method, that assigns a sample to predefined classes in three steps: (a) identification of genes that have strong correlations to parameters (predefined classes) of a training set of samples; (b) determination of an estimate of prediction error rates of training set by a leave-one-out cross-validation method; and (c) validation with an independent test set to obtain a true prediction error rate. In step 1, the Fisher exact test is used to identify genes by their degree of correlation to the predefined class (by user) of the training set of samples. Genes are then ranked by their predictive strength (negative natural log of p-value) that represents the probability of obtaining the observed number of samples from each class above or below the ideal pattern by chance. In step 2, samples from the training set are clustered using the k-NN method, where neighbors are identified by representing gene expression as vectors and placing samples according to the Euclidean distance. Each gene's discriminative ability is considered equally regardless of its value determined by Fisher's exact test (i.e., each classifier “votes” for a cohort and each vote is equal). After each gene evaluates the sample, the votes are summed to determine classification of the sample. Leave-one-out cross validation estimates the prediction error rate (or accuracy) by the systematic removal of one donor from the training set to use as a test sample. This process is repeated until all the donors have been “tested.” A p-value ratio cutoff of 0.2 was used in all discrimination analyses. A p-value ratio of 0.2 (equivalent to 1/5) indicates that the algorithm will make a prediction if the p-value (probability that the test sample is predicted as belonging to one class by chance) of the first best class is at least five times smaller than the p-value of the next best class. If the actual p-value ratio is less than the cutoff, a prediction will be made; if the ratio is higher, no prediction will be made. Setting the p-value cutoff to 1 will force the algorithm to always make a prediction but may result in more prediction errors. Thus, each gene will cast a vote for each sample in the dataset if the p-value from the predicted class is at least five times smaller than the other class. Given this, it is possible to have a tie-in class prediction (each predictor casts an equal vote), resulting in unclassified samples. In step 3, an independent sample set is evaluated, as in step 2, except for the leave-one-out cross-validation estimates.
For the comparison between PBMCs and cell subtypes, Affymetrix U133A and B GeneChip raw intensity data were scaled to a target intensity value of 500 (TGT) in Affymetrix Microarray Suite 5.0. Data were then imported into Genespring and analyzed without any further normalization steps.
RT-PCR.
RNA samples were DNase treated with TURBO DNA-free kit (Ambion), and total RNA for RT-PCR analysis was further amplified due to low yields of total RNA. 5 μg of each RNA sample was converted to cDNA using the High Capacity cDNA Archive kit (Applied Biosystems) in the PerkinElmer GeneAmp PCR System 9600. Quantitative PCR was performed on selected targets using pre-developed primers and probe TaqMan Gene Expression Assays (Applied Biosystems) on the ABI Prism 7700 Sequence Detection System. Expression results were calculated as the difference in cycle threshold relative to the median of four healthy volunteers for each target confirmed.
Online supplemental material.
Fig. S1 shows the differential gene expression profiles in PBMCs from healthy controls and SoJIA patients at three different stages of disease. Fig. S2 shows the analysis scheme that was followed to identify a SoJIA-specific signature. Fig. S3 shows the validation of the discriminative value of selected genes by RT-PCR. Table S1 shows the patient clinical data, and Table S2 shows a list of 873 genes distinguishing SoJIA patients from healthy controls. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070070/DC1.
Supplemental Material
Acknowledgments
We thank our patients and their parents/guardians for agreeing to participate in the study. We also thank Drs. Rolando Cimaz for critically reading the manuscript, Katherine Madson for her help recruiting and caring for these patients, and Daniel Kastner and Ivona Aksentijevich for kindly sharing microarray data with us.
This work was supported by Baylor Health Care System Foundation and the National Institutes of Health (R01 AR050770-01 to V. Pascual and CA78846 and U19A1057234-02 to J. Banchereau). J. Banchereau holds the Caruth Chair for Transplantation Immunology.
The authors have no conflicting financial interests.
Abbreviations used: CLIC-2, chloride intracellular channel 2; JIA, juvenile idiopathic arthritis; mDC, myeloid DC; pDC, plasmacytoid DC; SLE, systemic lupus erythematosus; SoJIA, systemic onset JIA.
F. Allantaz and D. Chaussabel contributed equally to this work.
References
- 1.Cassidy, J.T., and R.E. Petty. 2001. Textbook of Pediatric Rheumatology. W.B. Saunders, Philadelphia.
- 2.Wallace, C.A., and J.E. Levinson. 1991. Juvenile rheumatoid arthritis: outcome and treatment for the 1990s. Rheum. Dis. Clin. North Am. 17:891–905. [PubMed] [Google Scholar]
- 3.Woo, P., T.R. Southwood, A.M. Prieur, C.J. Dore, J. Grainger, J. David, C. Ryder, N. Hasson, A. Hall, and I. Lemelle. 2000. Randomized, placebo-controlled, crossover trial of low-dose oral methotrexate in children with extended oligoarticular or systemic arthritis. Arthritis Rheum. 43:1849–1857. [DOI] [PubMed] [Google Scholar]
- 4.Quartier, P., P. Taupin, F. Bourdeaut, I. Lemelle, P. Pillet, M. Bost, J. Sibilia, I. Kone-Paut, S. Gandon-Laloum, M. LeBideau, et al. 2003. Efficacy of etanercept for the treatment of juvenile idiopathic arthritis according to the onset type. Arthritis Rheum. 48:1093–1101. [DOI] [PubMed] [Google Scholar]
- 5.Ravelli, A., and A. Martini. 2003. Early predictors of outcome in juvenile idiopathic arthritis. Clin. Exp. Rheumatol. 21:S89–S93. [PubMed] [Google Scholar]
- 6.Modesto, C., P. Woo, J. Garcia-Consuegra, R. Merino, M. Garcia-Granero, C. Arnal, and A.M. Prieur. 2001. Systemic onset juvenile chronic arthritis, polyarticular pattern and hip involvement as markers for a bad prognosis. Clin. Exp. Rheumatol. 19:211–217. [PubMed] [Google Scholar]
- 7.Ravelli, A. 2004. Toward an understanding of the long-term outcome of juvenile idiopathic arthritis. Clin. Exp. Rheumatol. 22:271–275. [PubMed] [Google Scholar]
- 8.Spiegel, L.R., R. Schneider, B.A. Lang, N. Birdi, E.D. Silverman, R.M. Laxer, D. Stephens, and B.M. Feldman. 2000. Early predictors of poor functional outcome in systemic-onset juvenile rheumatoid arthritis: a multicenter cohort study. Arthritis Rheum. 43:2402–2409. [DOI] [PubMed] [Google Scholar]
- 9.Lomater, C., V. Gerloni, M. Gattinara, J. Mazzotti, R. Cimaz, and F. Fantini. 2000. Systemic onset juvenile idiopathic arthritis: a retrospective study of 80 consecutive patients followed for 10 years. J. Rheumatol. 27:491–496. [PubMed] [Google Scholar]
- 10.Bowyer, S.L., P.A. Roettcher, G.C. Higgins, B. Adams, L.K. Myers, C. Wallace, R. Rennebohm, T.L. Moore, P.H. Pepmueller, C. Spencer, et al. 2003. Health status of patients with juvenile rheumatoid arthritis at 1 and 5 years after diagnosis. J. Rheumatol. 30:394–400. [PubMed] [Google Scholar]
- 11.Pascual, V., F. Allantaz, E. Arce, M. Punaro, and J. Banchereau. 2005. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J. Exp. Med. 201:1479–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasques Godinho, F.M., M.J. Parreira Santos, and J. Canas da Silva. 2005. Refractory adult onset Still's disease successfully treated with anakinra. Ann. Rheum. Dis. 64:647–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, A.A., S.A. Leclercq, A. Yan, J.E. Homik, and C.A. Dinarello. 2005. Rapid responses to anakinra in patients with refractory adult-onset Still's disease. Arthritis Rheum. 52:1794–1803. [DOI] [PubMed] [Google Scholar]
- 14.Verbsky, J.W., and A.J. White. 2004. Effective use of the recombinant interleukin 1 receptor antagonist anakinra in therapy resistant systemic onset juvenile rheumatoid arthritis. J. Rheumatol. 31:2071–2075. [PubMed] [Google Scholar]
- 15.Aksentijevich, I., M. Nowak, M. Mallah, J.J. Chae, W.T. Watford, S.R. Hofmann, L. Stein, R. Russo, D. Goldsmith, P. Dent, et al. 2002. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 46:3340–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoham, N.G., M. Centola, E. Mansfield, K.M. Hull, G. Wood, C.A. Wise, and D.L. Kastner. 2003. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc. Natl. Acad. Sci. USA. 100:13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agostini, L., F. Martinon, K. Burns, M.F. McDermott, P.N. Hawkins, and J. Tschopp. 2004. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 20:319–325. [DOI] [PubMed] [Google Scholar]
- 18.Ramos, E., J.I. Arostegui, S. Campuzano, J. Rius, C. Bousono, and J. Yague. 2005. Positive clinical and biochemical responses to anakinra in a 3-yr-old patient with cryopyrin-associated periodic syndrome (CAPS). Rheumatology (Oxford). 44:1072–1073. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, H.M., S. Rosengren, D.L. Boyle, J.Y. Cho, J. Nayar, J.L. Mueller, J.P. Anderson, A.A. Wanderer, and G.S. Firestein. 2004. Prevention of cold-associated acute inflammation in familial cold autoinflammatory syndrome by interleukin-1 receptor antagonist. Lancet. 364:1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkins, P.N., H.J. Lachmann, E. Aganna, and M.F. McDermott. 2004. Spectrum of clinical features in Muckle-Wells syndrome and response to anakinra. Arthritis Rheum. 50:607–612. [DOI] [PubMed] [Google Scholar]
- 21.Lovell, D.J., S.L. Bowyer, and A.M. Solinger. 2005. Interleukin-1 blockade by anakinra improves clinical symptoms in patients with neonatal-onset multisystem inflammatory disease. Arthritis Rheum. 52:1283–1286. [DOI] [PubMed] [Google Scholar]
- 22.Wise, C.A., J.D. Gillum, C.E. Seidman, N.M. Lindor, R. Veile, S. Bashiardes, and M. Lovett. 2002. Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum. Mol. Genet. 11:961–969. [DOI] [PubMed] [Google Scholar]
- 23.Dierselhuis, M.P., J. Frenkel, N.M. Wulffraat, and J.J. Boelens. 2005. Anakinra for flares of pyogenic arthritis in PAPA syndrome. Rheumatology (Oxford). 44:406–408. [DOI] [PubMed] [Google Scholar]
- 24.Cassidy, J.T., J.E. Levinson, J.C. Bass, J. Baum, E.J. Brewer Jr., C.W. Fink, V. Hanson, J.C. Jacobs, A.T. Masi, J.G. Schaller, et al. 1986. A study of classification criteria for a diagnosis of juvenile rheumatoid arthritis. Arthritis Rheum. 29:274–281. [DOI] [PubMed] [Google Scholar]
- 25.Bennett, L., A.K. Palucka, E. Arce, V. Cantrell, J. Borvak, J. Banchereau, and V. Pascual. 2003. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J. Exp. Med. 197:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palucka, A.K., J.P. Blanck, L. Bennett, V. Pascual, and J. Banchereau. 2005. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc. Natl. Acad. Sci. USA. 102:3372–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson, H., P. Allen, and S.L. Peng. 2005. Inflammatory arthritis requires Foxo3a to prevent Fas ligand-induced neutrophil apoptosis. Nat. Med. 11:666–671. [DOI] [PubMed] [Google Scholar]
- 28.Chaussabel, D., W. Allman, A. Mejias, W. Chung, L. Bennett, O. Ramilo, V. Pascual, A.K. Palucka, and J. Banchereau. 2005. Analysis of significance patterns identifies ubiquitous and disease-specific gene-expression signatures in patient peripheral blood leukocytes. Ann. NY Acad. Sci. 1062:146–154. [DOI] [PubMed] [Google Scholar]
- 29.Dulhunty, A.F., P. Pouliquin, M. Coggan, P.W. Gage, and P.G. Board. 2005. A recently identified member of the glutathione transferase structural family modifies cardiac RyR2 substate activity, coupled gating and activation by Ca2+ and ATP. Biochem. J. 390:333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhoef, P.A., S.B. Kertesy, K. Lundberg, J.M. Kahlenberg, and G.R. Dubyak. 2005. Inhibitory effects of chloride on the activation of caspase-1, IL-1beta secretion, and cytolysis by the P2X7 receptor. J. Immunol. 175:7623–7634. [DOI] [PubMed] [Google Scholar]
- 31.Kim, K.T., K. Baird, J.Y. Ahn, P. Meltzer, M. Lilly, M. Levis, and D. Small. 2005. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood. 105:1759–1767. [DOI] [PubMed] [Google Scholar]
- 32.Ching, K.H., A.E. Kisailus, and P.D. Burbelo. 2005. The role of SPECs, small Cdc42-binding proteins, in F-actin accumulation at the immunological synapse. J. Biol. Chem. 280:23660–23667. [DOI] [PubMed] [Google Scholar]
- 33.Inoue, T., D. Hammaker, D.L. Boyle, and G.S. Firestein. 2005. Regulation of p38 MAPK by MAPK kinases 3 and 6 in fibroblast-like synoviocytes. J. Immunol. 174:4301–4306. [DOI] [PubMed] [Google Scholar]
- 34.Inoue, T., D.L. Boyle, M. Corr, D. Hammaker, R.J. Davis, R.A. Flavell, and G.S. Firestein. 2006. Mitogen-activated protein kinase kinase 3 is a pivotal pathway regulating p38 activation in inflammatory arthritis. Proc. Natl. Acad. Sci. USA. 103:5484–5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitney, A.R., M. Diehn, S.J. Popper, A.A. Alizadeh, J.C. Boldrick, D.A. Relman, and P.O. Brown. 2003. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA. 100:1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamero, A.M., and J.J. Oppenheim. 2006. IL-1 can act as number one. Immunity. 24:16–17. [DOI] [PubMed] [Google Scholar]
- 37.Miller, L.S., R.M. O'Connell, M.A. Gutierrez, E.M. Pietras, A. Shahangian, C.E. Gross, A. Thirumala, A.L. Cheung, G. Cheng, and R.L. Modlin. 2006. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity. 24:79–91. [DOI] [PubMed] [Google Scholar]
- 38.Woo, P. 2006. Systemic juvenile idiopathic arthritis: diagnosis, management, and outcome. Nat. Clin. Pract. Rheumatol. 2:28–34. [DOI] [PubMed] [Google Scholar]
- 39.Bhardwaj, N., L.L. Lau, S.M. Friedman, M.K. Crow, and R.M. Steinman. 1989. Interleukin 1 production during accessory cell-dependent mitogenesis of T lymphocytes. J. Exp. Med. 169:1121–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prieur, A.M., C. Bremard-Oury, C. Griscelli, and P. Mozziconacci. 1984. Prognosis of the systemic forms of juvenile chronic arthritis. Apropos of 100 cases. Arch. Fr. Pediatr. 41:91–97. [PubMed] [Google Scholar]
- 41.Ramilo, O., W. Allman, W. Chung, A. Mejias, M. Ardura, C. Glaser, K.M. Wittkowski, B. Piqueras, J. Banchereau, A.K. Palucka, and D. Chaussabel. 2007. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 109:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.