Abstract
Notch1 (N1) receptor signaling is essential and sufficient for T cell development, and recently developed in vitro culture systems point to members of the Delta family as being the physiological N1 ligands. We explored the ability of Delta1 (DL1) and DL4 to induce T cell lineage commitment and/or maturation in vitro and in vivo from bone marrow (BM) precursors conditionally gene targeted for N1 and/or N2. In vitro DL1 can trigger T cell lineage commitment via either N1 or N2. N1- or N2-mediated T cell lineage commitment can also occur in the spleen after short-term BM transplantation. However, N2–DL1–mediated signaling does not allow further T cell maturation beyond the CD25+ stage due to a lack of T cell receptor β expression. In contrast to DL1, DL4 induces and supports T cell commitment and maturation in vitro and in vivo exclusively via specific interaction with N1. Moreover, comparative binding studies show preferential interaction of DL4 with N1, whereas binding of DL1 to N1 is weak. Interestingly, preferential N1–DL4 binding reflects reduced dependence of this interaction on Lunatic fringe, a glycosyl transferase that generally enhances the avidity of Notch receptors for Delta ligands. Collectively, our results establish a hierarchy of Notch–Delta interactions in which N1–DL4 exhibits the greatest capacity to induce and support T cell development.
T cells, like other cells of the blood system, are derived from pluripotent hematopoietic stem cells (HSCs). The major site of T cell development is the thymus. Thus, descendants of HSCs migrate to the thymus, where they undergo a program of maturation, proliferation, and differentiation. They pass through a CD4−CD8− double-negative (DN) developmental stage, followed by a CD4+CD8+ double-positive (DP) stage, before undergoing positive or negative selection to generate single-positive (SP) CD4+ and CD8+ T cells that migrate to the periphery. The CD4−CD8− DN cells represent the most immature thymic subset that can be further subdivided into four developmental stages (DN1–4), based on their differential expression of CD44 and CD25, maturing from the CD44+CD25− (DN1) to the CD44+CD25+ (DN2) to the CD44−CD25+ (DN3) to the CD44−CD25− (DN4) stages (1–3). Many different signaling pathways have been shown to be involved in T lymphocyte development. One of these pathways is the Notch cascade, which has received a lot of attention in recent years because of its involvement in T lineage commitment, T cell maturation, and peripheral T cell function (4, 5). Notch proteins compose a family of four transmembrane receptors that influence cell fate decisions and differentiation processes in many different organisms (6). Notch signaling is triggered upon binding of ligands of the Jagged and Delta family. This leads to a cascade of proteolytic cleavages that release the intracellular cytoplasmic domain of Notch receptors, which subsequently translocates to the nucleus, where it binds to the RBP-J transcription factor and thereby activates transcription. Notch signaling itself can be regulated by several modulators, such as the family of Fringe proteins, which are glycosyl transferases that add N-acetylglucosamine to certain epidermal growth factor–like repeats of Notch receptors, promoting Notch signaling in response to Delta ligands and inhibiting Jagged-mediated Notch signaling (7).
The best-established role for Notch signaling in the hematopoietic system is the essential function of Notch1 (N1) in T cell fate specification. Conditional inactivation of the N1 (8, 9) or RBP-J (10) genes in adult BM progenitors results in B cell development within the thymus at the expense of T cell lineage commitment, suggesting that N1/RBP-J–mediated signaling is important to induce T cell development and to simultaneously block B cell development. Although multiple Notch receptors such as N1, N2, and N3—as well as the ligands Jagged1, Jagged2, Delta1 (DL1), and DL4—are expressed on thymocytes and/or thymic epithelium (11–18), T lineage commitment appears to be mediated via the N1 receptor in a nonredundant manner. This is consistent with the finding that conditional inactivation of the N2 gene does not affect T cell development but is instead necessary for marginal zone B cell (MZB cell) specification (19). Moreover, N3 gene–targeted mice do not exhibit any hematopoietic phenotype (20). Further support for the essential role of Notch signaling in T cell lineage commitment is derived from gain-of-function studies, as overexpression of a constitutively active form of N1 (21, 22) or DL4 (13, 23, 24) induces ectopic T cell development in the BM and simultaneously blocks B cell development. Thus, these reciprocal loss- and gain-of-function studies indicate that N1 signaling is necessary and sufficient for T cell lineage commitment.
An additional nonredundant function of N1 during thymocyte maturation was revealed by conditional inactivation of the N1 gene in immature thymocytes. N1 deficiency in DN thymocytes leads to a partial block of αβ T cell development at the pre-TCR checkpoint because of defective V to DJβ rearrangement (25). Although N1 seems to be a key player during T lineage commitment and T cell maturation, several issues are still controversial or unknown. For example, the expression of multiple Notch ligands on thymic epithelial cells leads to the question of which ligand triggers the physiological N1 signal for T lineage commitment and/or maturation. Ligands of the Jagged family (Jagged1 and Jagged2) can be excluded as being essential during these processes, because conditional inactivation of Jagged1 (26) does not perturb hematopoiesis and Jagged2-deficient mice show only a minor decrease in γδ T cells, whereas αβ T cell development appears normal (27). Thus, members of the Delta-like family seem to be the crucial ligands, because expression of DL1 or DL4 on stromal cells can induce T cell development of human or mouse hematopoietic progenitors (28–30). Interestingly, conditional inactivation of the DL1 gene in hematopoietic cells leads to the loss of MZB cells, indicating that DL1 signals via N2 to specify this subclass of splenic B cells (30). Surprisingly, loss of DL1, even in thymic epithelium, does not perturb T cell development, indicating that DL1 is also dispensable for T cell lineage commitment or T cell maturation in vivo (19, 30). Because DL4-expressing stromal cells can also induce T cell development in vitro (18, 30), it is conceivable that loss of DL1 function in vivo is compensated by DL4.
In this paper, we explore the ability of DL1 and DL4 to induce T cell lineage commitment and/or to influence T cell maturation in vitro and in vivo via interactions with N1 and/or N2. Our results show that DL1 and DL4 exhibit different Notch receptor specificities and that T cell fate specification is mediated by specific Notch receptor–ligand interactions.
RESULTS
N2 compensates for the loss of N1 during T cell commitment in vitro but not in vivo
To further characterize the Notch-dependent events of T cell development, we examined in vivo versus in vitro T lineage commitment. For the in vivo analysis, we set up mixed BM chimeras using BM cells of conditional gene-targeted mice in which either the N1 or N2 genes alone or both N1 and N2 can be inactivated simultaneously in HSCs (using the IFN-α–responsive Mx-Cre system). Therefore, CD45.1+ WT lethally irradiated mice were injected with either CD45.2+ control (N1lox/lox; Ctrl), N1-deficient (N1−/−), N2-deficient (N2−/−), or N1N2–double-deficient (N1N2−/−) BM progenitors. Thymic T cell development was analyzed 8 wk after transplantation (Fig. 1 A). As previously described, inactivation of the N1 gene in BM progenitors results in a block at or before the earliest intrathymic precursor stage (8). Immature B cells develop in the thymus from incoming N1−/− BM progenitors, demonstrating that N1 signaling is essential for T lineage commitment in vivo (Fig. 1 A) (9). N2−/− BM progenitors reconstituted the T cell lineage as efficiently as BM cells derived from control animals. No ectopic B cell development was observed in the thymus of N2−/− BM chimeras. The only detectable hematopoietic phenotype caused by the inactivation of N2 was the loss of MZB cells in the spleen (unpublished data). In contrast, chimeric mice reconstituted with BM progenitors double deficient for both N1 and N2 recapitulated the phenotype of the N1−/− BM chimeras, suggesting that T lineage commitment in vivo is exclusively dependent on N1 signaling.
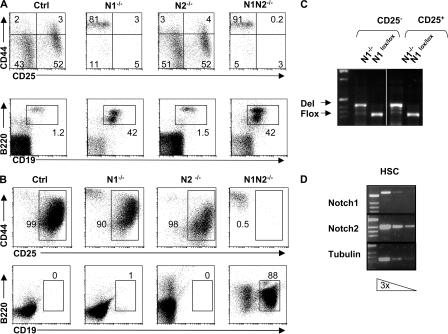
Figure 1.
N2 signaling is sufficient to induce T lineage commitment in vitro but not in vivo. (A) Mixed BM chimeric mice were analyzed 8 wk after reconstitution with a 1:2 mixture of WT (CD45.1+) and Ctrl (N1lox/lox), N1−/−, N2−/−, or N1N2−/− (CD45.2+) BM-derived populations. Representative FACS analysis of thymocytes stained with anti-CD117, -CD44, and -CD25 antibodies after gating on donor (CD45.2+)-derived lineage-negative cells (top). Representative FACS analysis of thymocytes stained with anti-B220 and -CD19 antibodies after gating on donor (CD45.2+)-derived lineage-negative cells (bottom). Representative FACS profiles are derived from experiments in which five mice of each genotype were analyzed. (B) BM KLS cells were sorted from Ctrl, N1−/−, N2−/−, and N1N2−/− mice and cultured on OP9-DL1 cells for 10 d (top) and 18 d (bottom). Cells from these cultures were analyzed for the expression of CD44 and CD25 (top) and for the presence of B220+CD19+ B cells (bottom). Representative FACS profiles are derived from four individual experiments. (C) Deletion PCR analysis for the N1 gene was performed on genomic DNA from sorted CD25− (corresponding to DN1) and CD25+ (corresponding to DN2/DN3) cells derived from N1−/− and Ctrl animals cultured for 10 d on OP9-DL1. (D) Semiquantitative RT-PCR for the expression of N1, N2, and tubulin was performed on sorted BM KLS cells. Three serial dilutions (threefold) of template RNA are shown for the indicated genes.
To further characterize the N1-dependent events of T lineage commitment in vitro, we have made use of the OP9-DL1 culture system (29) in which purified (CD117+lin−Sca1+; KLS) HSCs from either Ctrl, N1−/−, N2−/−, or N1N2−/− BM progenitors were sorted and cultured on DL1-expressing OP9 stromal cells. Surprisingly, after 10 d of culture, N1−/− HSCs principally gave rise to DN2 (CD44+CD25+) and DN3 (CD44−CD25+) T cell progenitors (Fig. 1 B). No difference was observed in the ability of Ctrl or N1−/− HSCs to differentiate into immature thymocytes. To rule out the possibility that these OP9-DL1–generated DN2 and DN3 progenitor cells were derived from precursor cells that had escaped N1 deletion, both subsets were sorted and analyzed by PCR for the successful inactivation of the N1 gene. The PCR analysis shows the expected bands characterizing the floxed and the inactivated N1 alleles of the sorted cells, respectively, confirming that the OP9-DL1–generated DN2 and DN3 cells were indeed the progeny of N1−/− HSCs (Fig. 1 C).
Because N2 is expressed together with N1 on HSCs (Fig. 1 D), it is conceivable that N2 is able to compensate for the loss of N1 function during T lineage commitment when N1−/− HSCs are cultured on DL1-expressing OP9 cells. To test this hypothesis, we sorted and cultured N2−/− and N1N2−/− HSCs on DL1-expressing OP9 cells. After 10 d of OP9-DL1 culture, N2−/− HSCs gave rise to immature DN1–3 T cell progenitors similar to Ctrl and N1−/− HSCs (Fig. 1 B). In contrast, N1N2−/− HSCs do not develop into T cell progenitors; instead, they exhibit a developmental block at the putative DN1 stage characterized by the accumulation of CD44+CD25− cells. This phenotype is very reminiscent of the defect observed in vivo in inducible N1−/− mice, where CD44+CD25− cells accumulated in the thymus and were identified as B220+CD19+ B cells (Fig. 1 A) (8). Therefore, Ctrl, N1−/−, N2−/−, and N1N2−/− HSCs were assessed for their ability to develop into B cells on OP9-DL1–expressing stromal cells. Only HSCs derived from N1N2−/− HSCs developed into B cells after 18 d of culture, whereas Ctrl, N1−/−, or N2−/− cells did not. These data confirm the hypothesis that DL1-mediated N2 signaling can compensate for the loss of N1 function during T lineage commitment in vitro and that the presence of either N1 or N2 alone is sufficient to block B cell development.
N2 cannot compensate for the loss of N1 function during T cell maturation
Notch signaling is not only essential for T lineage commitment but is also continuously required for the successful differentiation of all DN thymocyte subsets into CD4+CD8+ DP cells (14). Because N2 can instruct N1−/− HSCs to adopt a T cell fate on OP9-DL1–expressing stromal cells, it is conceivable that N2 signaling would be sufficient to allow the subsequent DN to DP transition. To this end, Ctrl and N1−/− HSCs were cultured for 28 d on OP9-DL1 stromal cells and subsequently analyzed for the development of DP T cells. Although Ctrl HSCs differentiated very efficiently into DP T cells, >90% of the N1−/− cells appeared to be blocked in the DN compartment (Fig. 2 A).
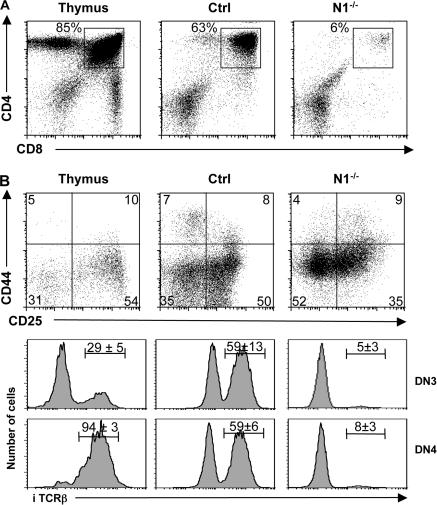
Figure 2.
N2 cannot compensate for the loss of N1 function during T cell maturation in vitro. (A) KLS cells from Ctrl and induced N1−/− mice were sorted and cultured on OP-DL1 cells for 20 d. A representative flow cytometric analysis of CD4 versus CD8 of WT thymocytes, and Ctrl and N1−/− KLS cells cultured on OP9-DL1 are shown. (B) Indicated cells were electronically gated on lineage-negative DN thymocytes and analyzed for the expression of CD44 and CD25. Representative histograms for intracellular TCRβ (iTCRβ) expression on DN3 and DN4 thymocytes derived from Ctrl and N1−/− KLS cells 20 d after culture on OP9-DL1 are shown. The numbers above the bars indicate the percentage ± SD of iTCRβ+ cells (n = 4 for WT thymocytes and in vitro culture experiments).
These data show that N2 signaling, although sufficient for T cell commitment of BM HSCs, is not sufficient for T cell maturation. Impaired DN to DP transition has also been observed in mice in which the N1 gene was inactivated in immature thymocytes using the lck-Cre transgene (25). In these mice, N1 deficiency leads to a partial block of αβ T cell development at the pre-TCR checkpoint because of inhibition of VDJβ rearrangement. This partial block is characterized by a substantial decrease in the proportion of N1−/− DN3 and DN4 cells expressing intracellular TCRβ protein (25). To investigate whether the block observed in the in vitro culture system at the DN to DP transition might also be caused by inefficient expression of a TCRβ chain, intracellular TCRβ staining was performed on WT, DN3, or DN4 thymocytes or on in vitro–generated DN T cell progenitors derived from Ctrl or N1−/− HSCs. Approximately 60% of the in vitro–generated DN3 and DN4 T cell progenitors derived from Ctrl HSCs have an in-frame TCRβ rearrangement and, thus, stained positive for intracellular TCRβ, compared with 29% of DN3 and 94% of DN4 thymocytes in vivo (Fig. 2 B). However, only 5 and 8% of the DN3 and DN4 cells, respectively, derived from N1−/− HSCs were icTCRβ+. These results show that DL1-mediated N2 signaling is not sufficient to allow differentiation of DN immature cells into DP T cell progenitors in the absence of N1 because of impaired TCRβ rearrangement and/or expression.
N2 mediates T lineage commitment in vivo in the absence of N1 signaling at extrathymic sites after BM transplantation
Although N1 is the key receptor for T lineage commitment in the thymus, our in vitro data raise the possibility that, under certain conditions (when encountering the DL1 ligand), the N2 receptor might be able to induce T lineage commitment at extrathymic sites in vivo.
Recent studies by Maillard et al. (31), Lancrin et al. (32), and Arcangeli et al. (33) showed that early T cell development occurs in the spleen and LNs after BM transplantation. This pool of splenic T cell progenitors can efficiently contribute to donor-derived thymopoiesis by migrating from the spleen to the thymus (31), where they complete T cell maturation. The generation of these splenic T cell progenitors after BM transplantation is Notch signaling dependent (31), because it is blocked by the expression of a dominant-negative form of the Notch coactivator mastermind-like 1 (31). However, it is not clear whether these extrathymically derived T cell progenitors are generated in a N1- and/or N2-dependent manner. To address this question, we transplanted CD45.2+ Ctrl, N1−/−, N2−/−, and N1N2−/− BM cells into lethally irradiated CD45.1+ C57BL/6 recipients. Donor-derived lin− cells in the spleen were analyzed 12 d after BM transplantation by staining for Thy1.2, CD44, and CD25. As previously reported, Thy1.2 and CD44 staining identified two populations within the lin− donor-derived cells in mice receiving Ctrl BM cells (32, 33). The CD44+Thy1.2− population has previously been shown to have multilineage potential, whereas the CD44lo/−Thy1.2+ population is T lineage restricted (Fig. 3) (33). The Thy1.2+ cells appear to be heterogeneous for the expression of CD25, as ∼50% express this marker, which normally defines the DN2 and DN3 subsets of immature thymocytes. A similar population of splenic Thy1.2+ T cell progenitors was identified in hosts receiving Ctrl, N1−/−, and N2−/− BM. In contrast, no Thy1.2+ cells were observed in the lin− donor-derived population of hosts receiving N1N2−/− BM (Fig. 3). These results demonstrate that splenic T cell progenitors can be generated in the absence of N1 after BM transplantation in a N2-dependent manner. Thus, either N2 or N1 signaling is sufficient for T lineage commitment in the spleen after BM transplantation.
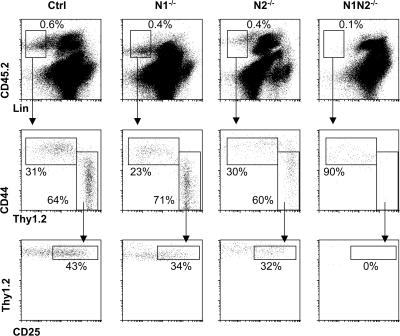
Figure 3.
N2 is sufficient to specify T lineage progenitors in the spleen after BM transplantation. CD45.2+ Ctrl, N1−/−, N2−/−, or N1N2−/− BM cells were injected into lethally irradiated CD45.1+ hosts. The spleens of host mice were analyzed 12 d after BM transplantation. Representative flow cytometric analyses of donor-derived lineage-negative cells for the expression of CD44 and Thy1.2, and Thy1.2 and CD25, respectively, are shown. Data are representative of four independent experiments.
DL1 and DL4 ligands exhibit different Notch receptor specificities
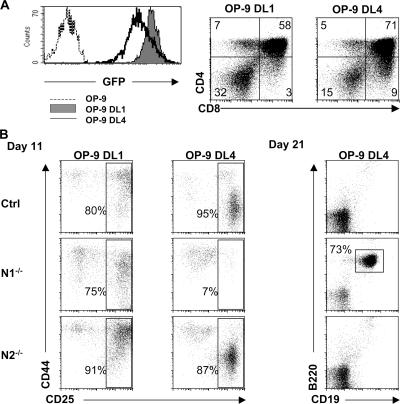
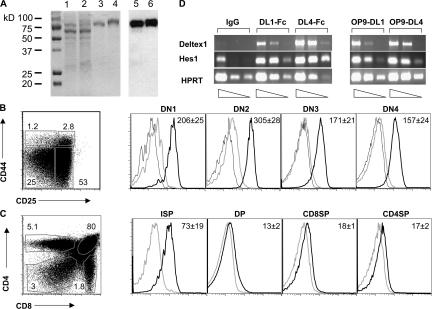
The simplest hypothesis to explain the discrepancy in the ability of N2 to compensate for the loss of N1 during T lineage commitment in vitro but not in vivo is that the N2 gene, although being expressed on HSCs, is not expressed in the earliest thymocyte progenitors. To test this hypothesis, semiquantitative RT-PCR was performed on all DN subsets derived from either BM HSCs that have been cultured on OP9-DL1 cells or from WT thymocytes. As shown in Fig. 4, both N1 and N2 were expressed in all DN subsets (even in DN1, CD117+CD44+CD25− subsets), irrespective of whether they were generated in vitro or were derived from thymocytes in vivo. Furthermore, no noticeable differences were observed in the expression levels of both genes in the different DN subsets (Fig. 4). Thus, the hypothesis that the absence of N2 expression in early thymocyte progenitors accounts for the differential outcome of T lineage commitment in vivo versus in vitro is unlikely. However, an alternative explanation for this discrepancy is that the outcome of Notch signaling might be dependent on the specificity and/or avidity of certain ligand–receptor interactions. Although DL1 is able to interact with N1 and N2 to trigger T lineage commitment in vitro, it is conceivable that other Notch ligands may also trigger T lineage commitment, but only upon selective interaction with one specific Notch receptor. An attractive alternative ligand for N1 is DL4, a close homologue of DL1, which has also been shown to trigger T lineage commitment of WT precursors in vitro when expressed on OP9 cells (30). To test whether DL1 and DL4 can mediate T cell development equally efficiently in vitro, sorted WT BM HSCs were cultured on DL1- or DL4-expressing OP9 cells and analyzed side by side. Although the expression level of DL4 was slightly lower than DL1 on OP9 cells (as judged by GFP expression; Fig. 5 A, left), WT HSCs developed very efficiently into DP thymocytes in both cases within 30 d of culture (Fig. 5 A, right). To investigate whether DL1 and DL4 exhibit different Notch receptor specificities, Ctrl, N2−/−, and N1−/− HSCs were cultured side by side on either DL1- or DL4-expressing OP9 cells. After 11 d of culture, Ctrl HSCs cultured on either DL1 or DL4 progressed to the DN2/DN3 stage, although the progression appeared to be slightly more rapid on OP9-DL4–expressing cells. Whereas all three genotypes developed into DN1, DN2, and DN3 T cell progenitors on DL1-expressing OP9 cells, only Ctrl and N2−/− HSCs, but not N1−/− HSCs, developed into DN1, DN2, and DN3 T cell progenitors when cultured on OP9-DL4 cells (Fig. 5 B). Within the same time frame, N1−/− HSCs could only differentiate into a putative DN1 population (CD44+CD25−) on OP9-DL4 cells. These data indicate that N1−/− HSCs either exhibit a developmental block at the DN1 to DN2 transition or that they may have adopted a B cell fate similar to the phenotype observed after inducible inactivation of N1 in BM progenitors in vivo (Fig. 1 A) (8, 9). Therefore, the different OP9-DL4 cultures were stained with antibodies to the pan–B cell markers B220 and CD19. As shown in Fig. 5 B (right), B220+CD19+ B cells were only observed when N1−/− but not Ctrl or N2−/− BM HSCs were cultured on OP9-DL4 cells. These results demonstrate that DL4 and DL1 are not equivalent in their ability to trigger T lineage commitment, as DL4 can only induce T lineage commitment of BM HSCs that express N1. Moreover, DL4-mediated N2 signaling is not sufficient for T cell commitment nor is it sufficient for the inhibition of B cell development in vitro, suggesting that DL4 signals specifically through the N1 receptor, whereas DL1 can signal through both N1 and N2. To investigate whether DL1 and DL4 exhibit different binding avidities for N1 and/or N2, DL1- and DL4-IgG fusion proteins were generated (Fig. 6 A) and assessed for their ability to bind to Notch receptors expressed on thymocytes. As shown in Fig. 6 B, DL4-IgG fusion proteins bind immature DN thymocytes very efficiently. Binding of DL4-IgG is already observed in the DN1 subset, peaks at the DN2 subset, gradually declines through the DN3, DN4, and immature SP (ISP) subsets, and is virtually absent in more mature DP and CD4 and CD8 SP thymocytes (Fig. 6 C). Surprisingly, DL1-IgG fusion proteins did not stain thymocytes above background levels of the IgG isotype control (unpublished data), with the exception of the DN1 and DN2 subsets, which stained weakly above background. These results demonstrate that the DL4 fusion protein has a considerably higher binding avidity to Notch receptors present on immature thymocytes compared with DL1.
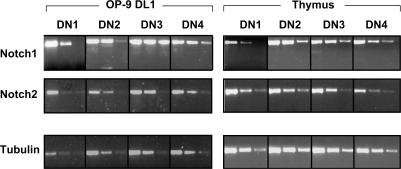
Figure 4.
Expression of N1 and N2 in immature DN thymocytes. cDNA was prepared from sorted cells cultured on OP9-DL1 cells for 16 d (corresponding to the DN1–4 subsets), and WT DN1–4 thymocyte subsets. Transcripts of N1 and N2 were analyzed by semiquantitative RT-PCR of threefold dilutions of the cDNA. The cDNA input was normalized according to the expression of the control tubulin gene.
Figure 5.
Comparison of DL1- and DL4-mediated T cell development in vitro. (A) Histograms show flow cytometric analyses of GFP expression of uninfected OP9-cells (dashed line) and DL1 (shaded histogram) and DL4 (continuous line) retrovirally transduced OP9 cells. Ctrl BM HSCs were sorted and cultured side by side on OP9-DL1 and -DL4 cells and anaylzed by flow cytometry for the expression of CD4 and CD8 30 d after culture. (B) Sorted BM HSCs derived from either Ctrl, N1−/−, or N2−/− mice were cultured for the indicated times on either OP9-DL1 or -DL4 cells and subsequently analyzed by flow cytometry for the expression of CD44 and CD25 (electronically gated on lineage-negative cells) and the presence of B220+CD19+ B cells. Data are representative of four independent experiments.
Figure 6.
Binding of purified DL1- and DL4-IgG fusion proteins to thymocytes. (A) DL1- (lanes 1 and 3) and DL4-IgG fusion proteins (lanes 2 and 4) before (lanes 1 and 2) and after (lanes 3 and 4) purification over a protein A column were stained with Coommassie blue (left). A Western blot analysis of DL1- (lane 5) and DL4-IgG (lane 6) fusion proteins using an anti–human IgG–horseradish peroxidase–conjugated antibody is also shown (right). (B) The purified DL1- and DL4-IgG fusion proteins were used to stain immature WT thymocytes. Thymocytes were stained with lineage cocktail and anti-CD117, -CD44, and -CD25 antibodies together with DL1- or DL4-IgG fusion proteins. Representative histograms show the staining of DL1- (gray line) and DL4-IgG (bold line) fusion proteins or IgG isotype control (continuous line) gated on the DN1 (CD117+CD44+CD25−), DN2 (CD117+CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44−CD25−) thymocyte subpopulations. (C) Representative histograms showing staining of DL1- (gray line) and DL4-IgG (bold line) fusion proteins gated on ISP (CD8+TCRβ−), DP (CD4+CD8+), CD4SP (CD4+TCRβ+), and CD8SP (CD8+TCRβ+) thymocytes. In the more mature thymocyte subsets (ISP, DP, and SP), DL1-IgG staining was indistinguishable from the IgG isotype control, which is therefore not shown in C. Data are representative of four independent experiments, and numbers within the histograms indicate means ± SD of the mean fluorescence intensity for DL4. (D) Semiquantitative RT-PCR for the Notch target genes Deltex1 and Hes1. cDNA was prepared from DN thymocytes, either cultured for 20 h on IgG-, (DL1-IgG) DL1-Fc–, and DL4-Fc–coated plastic dishes or on OP9-DL1 or -DL4 cells. Transcripts of Deltex1 and Hes1 were analyzed by semiquantitative RT-PCR of threefold dilutions of the cDNA. The cDNA input was normalized according to the expression of the control HPRT gene.
To investigate whether differences in the binding avidity of DL1- and DL4-IgG fusion proteins translate into differential Notch target gene induction, immature DN thymocytes were cultured on either DL1- or DL4-IgG–coated plastic dishes or on DL1- or DL4-expressing OP9 stromal cells. Expression of the Notch target genes Deltex1 and Hes1 was subsequently analyzed by semiquantitative RT-PCR. Similar to the hierarchy observed in binding assays, induction of Deltex1 gene expression was stronger when Notch signaling was triggered by DL4 compared with DL1. However, differences in Hes1 gene expression were less pronounced or, as in the OP9 culture system, not observed (Fig. 6 D). These results indicate that differential Notch–Delta binding avidity correlates in some cases with differences at the level of gene expression. However, this correlation appears to be target gene dependent, suggesting that different target genes respond to different threshold levels of Notch signaling.
Because the binding assays were performed with WT thymocytes, we were unable to distinguish binding of DL4-IgG to the N1 and/or N2 receptor. Our results presented in Fig. 5 show that DL4, in contrast to DL1, cannot induce T lineage commitment via N2. This result is compatible with two hypotheses: either DL4 cannot bind efficiently to N2 or, alternatively, DL4 can bind N2 but cannot transmit a signal via the N2 receptor. To distinguish between these two possibilities, we examined the binding efficiency of DL1- and DL4-IgG fusion proteins to 293T cells transiently expressing either N1- or N2–enhanced GFP (EGFP) fusion proteins. As shown in Fig. 7 (B and C, top), DL4-IgG fusion proteins bind N1 very efficiently, whereas binding to the N2 receptor is not detectable above background. Interestingly, DL1-IgG fusion proteins bind N1 weakly but do not bind N2 above levels of the IgG isotype control.
Figure 7.
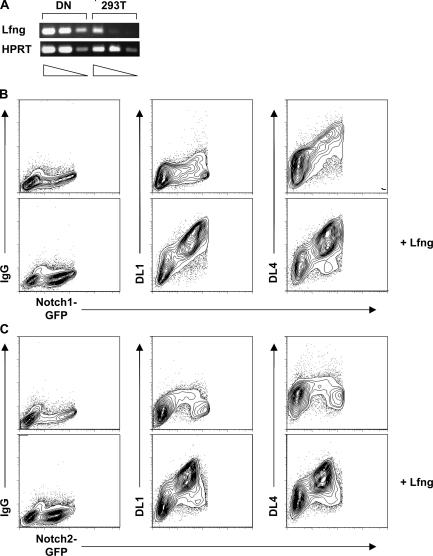
Binding of DL1- and DL4-IgG fusion proteins to N1 and N2. (A) Semiquantitative RT-PCR for the Lfng gene. cDNAs were prepared from DN thymocytes and 293T cells. Transcripts were analyzed by semiquantitative RT-PCR of fivefold dilutions of the cDNA. The cDNA input was normalized according to the expression of the control HPRT gene. 293T cells were transiently transfected with N1- (B) or N2-EGFP (C) together with or without Lfng expression vectors and stained 48 h after transfection with either human IgG1 isotype control or DL1- or DL4-IgG fusion proteins. Data are representative FACS profiles of four independent experiments. Extremely high EGFP-expressing cells were gated out as the fusion proteins were trapped inside the cells.
Modulation of Notch–Delta binding avidity by Lunatic fringe (Lfng)
A recent report suggests that the sensitivity of Notch receptors to DL ligands in thymocytes is regulated by the glycosyltransferase Lfng (34). Although only N1–DL4 binding can be detected in transiently transfected 293T cells (Fig. 7 B, top), it is possible that this situation reflects the fact that 293T cells express very low levels of Lfng as measured by semiquantitative RT-PCR (Fig. 7 A). To investigate directly whether the avidity of Notch–Delta binding might be dependent on Lfng expression, 293T cells transiently expressing either N1 or N2 were cotransfected with an expression plasmid encoding Lfng and subsequently analyzed in binding assays using DL1- and DL4-IgG fusion proteins. Interestingly, overexpression of Lfng in 293T cells allows DL1 to bind efficiently to N1 and N2. Moreover, DL4-Fc can now also bind N2 (Fig. 7 B and C, bottom). These results directly demonstrate that the binding avidity of DL ligands to Notch receptors is Lfng dependent, with the possible exception of the DL4–N1 interaction. They furthermore highlight the possibility that Notch signaling might be regulated in an important way at the level of Lfng expression.
Enforced expression of DL4 but not DL1 induces efficient T cell development in vivo
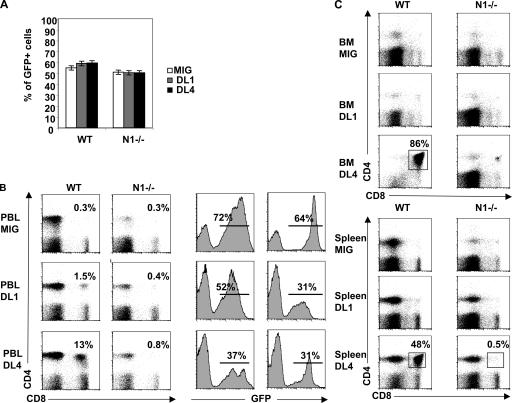
In vitro, DL4 is unable to induce T lineage commitment in the absence of a functional N1 receptor, suggesting that DL4 must specifically interact with N1 to specify the T lineage. To exclude that this observation is caused by a peculiarity of our in vitro culture system, we investigated the ability of DL1 and DL4 to induce T cell development in vivo in the presence and absence of N1. Previous studies demonstrated that retroviral overexpression of DL4 in hematopoietic cells is sufficient to promote thymus-independent T cell development to the DP stage in vivo (13, 23, 24). We therefore transduced CD45.2+ Ctrl and N1−/− BM cells with a retrovirus expressing either GFP alone (MIG), or DL1 or DL4 together with GFP, and subsequently transplanted these cells into lethally irradiated CD45.1+ C57BL/6 mice. The BM transduction efficiency of MIG and MIG expressing either DL1 or DL4 virus (based on GFP expression) was between 55 and 60% for both Ctrl and N1−/− BM cells (Fig. 8 A). Reconstituted hosts were analyzed 9 wk after transplantation for the presence of GFP+ donor-derived cells in PBLs. 72% of Ctrl PBLs and 64% of N1−/− PBLs in host mice that were transplanted with MIG-transduced BM cells were GFP+. Comparable percentages of PBLs were GFP+ in hosts receiving either DL1- or DL4-expressing Ctrl and N1−/− BM cells, indicating that the relative number of virus-expressing Ctrl and N1−/− donor cells was comparable even 9 wk after transplantation (Fig. 8 B, right). Only forced expression of DL4 but not DL1 resulted in the efficient development of DP T cells (Fig. 8, B–D). DL4-induced DP T cells were exclusively found in the PBLs (13%), BM (86%), and spleen (48%) of Ctrl but not of N1−/− chimeras, suggesting that enforced DL4 expression can only induce T cell development of N1-expressing progenitors in vivo. These results confirm our in vitro results using the DL4-expressing OP9 cells.
Figure 8.
Comparative in vivo analysis of DL1 and DL4 for their ability to promote ectopic T cell development. (A) Bar diagrams show percentages of GFP+ Ctrl and N1−/− BM cells after transduction with the control virus (MIG) and virus expressing DL1 and DL4. (B) Histograms show the percentage of GFP+ cells within total PBLs in host mice 9 wk after BM transplantation. Flow cytometric analysis for the presence of CD4- and CD8-expressing T cells was performed on PBLs (B) and BM and spleen (C) of host animals that were transplanted with WT and N1−/− BM cells expressing the indicated ligands.
DISCUSSION
The data presented in this study provide compelling evidence for a hierarchy of Notch–Delta interactions promoting T cell lineage commitment and maturation. Using the well-characterized OP9 stromal cell culture system, we unexpectedly found that DL1 can trigger T cell lineage commitment (to the DN CD25+ stage) via either N1 or N2 receptors expressed on BM precursors. In contrast, DL4 could only induce T cell lineage commitment via N1. Concomitant with T cell lineage commitment, inhibition of B cell development was observed for N1–DL1, N1–DL4, and N2–DL1 receptor–ligand pairs but not for N2–DL4. Collectively, our data demonstrate novel aspects of both the specificity and redundancy of Notch receptor–ligand interactions in the induction of T cell lineage commitment on OP9 stromal cells. Furthermore, they highlight the fact that N2 is unable to promote thymic T cell lineage commitment in vivo despite its ability to induce T cell commitment in vitro via DL1. This unexpected ability of N2 to substitute for N1 in the promotion of T cell lineage commitment was also observed in vivo in a short-term BM transplantation model, originally derived to quantitate pluripotential stem cells (35). In this system, spleen colonies in lethally irradiated recipients derived from single BM stem cells contain multiple hematopoietic lineages, including committed T cell precursors expressing Thy-1 and CD25 (32, 33). Development of these extrathymic T cell precursors was recently shown to depend upon Notch signaling, because it is inhibited by a dominant-negative form of the Notch coactivator mastermind-like 1 (31). Our data using conditional Notch knockout BM precursors clearly demonstrate that expression of either N1 or N2 is sufficient to promote T cell lineage commitment in the spleen after transplantation. The relevant Notch ligands responsible for T cell commitment in this system have not been identified. Nevertheless, it is clear that DL1 is expressed on both B cells and DCs in the spleen (30), and DL1 is the nonredundant ligand responsible for N2-mediated MZB cell fate specification in that organ (30). By analogy with the OP9 system, it is thus tempting to speculate that N2–DL1 interactions account for the ability of N2 to substitute for N1 in promoting extrathymic T cell lineage commitment after short-term BM transplantation.
Notch–Delta interactions are not only required for T cell lineage commitment but also for further maturation of DN CD25+ immature thymocytes to the DP stage both in vivo (25) and in vitro (14). A critical aspect of this maturation process is productive VDJ rearrangement of the TCRβ locus, leading to expression of a TCRβ protein and functional pre-TCR. In the OP9 system, it is known that both DL1 and DL4 can promote TCRβ rearrangement and progression to the DP stage when WT BM precursors are plated (18). However, DL1 was unable to promote the further maturation of DN CD25+ precursors that had developed via N2 (i.e., in the absence of N1). Furthermore, this defect in the generation of DP thymocytes was accompanied by a failure to express TCRβ protein in DN CD25+ cells. Collectively, these data indicate that N2–DL1 interactions, though sufficient to promote T cell lineage commitment in vitro, are unable to induce subsequent T cell maturation. Thus, it is possible that N2–DL1 interactions in the OP9 system are of lower avidity than N1–DL1 and, especially, N1–DL4 interactions, as further suggested by direct binding assays of DL1 and DL4 fusion proteins on N1- or N2-transfected 293T cells. Nevertheless, N2–DL1 interactions are readily detected in the presence of high levels of Lfng in transfected 293T cells, raising the possibility that the failure of N2–DL1 to induce T cell maturation in vitro could be explained by limiting concentrations of Lfng in thymic progenitors. Alternatively, N2 may signal less efficiently than N1 because of its weaker transactivation domain (4).
An even more stringent requirement for Notch–Delta interactions in T cell maturation was observed in an in vivo model where DL1 and DL4 were retrovirally transduced in WT and N1−/− BM precursors that were subsequently used to reconstitute lethally irradiated hosts. In agreement with several other reports (13, 23, 24), large numbers of DP cells developed after 9 wk in the BM, spleen, and PBLs of mice reconstituted with DL4-expressing WT BM precursors. In contrast, DL1-expressing WT BM precursors did not generate detectable numbers of DP cells. Importantly, DL4-induced generation of DP cells was totally dependent on N1, because it did not occur when reconstitution was performed with DL4-transduced N1−/− BM precursors. These results point to a highly specific N1–DL4 interaction as being essential for extrathymic T cell maturation to the DP stage in vivo when Delta expression is restricted to hematopoietic cells. The unique N1–DL4 specificity in this system could be related to the fact that the N1–DL4 interaction is less dependent on Lfng than other Notch–Delta interactions. According to this scenario, putative low levels of Lfng in HSCs could restrict their ability to undergo T cell maturation in the BM unless they encounter DL4 on hematopoietic cells. This stringent requirement may be overcome in the OP9-DL1 system, where high levels of expression of DL1 and/or other co-stimulatory properties of OP9 stromal cells may compensate for the putatively weaker N1–DL1 interaction.
The hierarchal nature of Notch–Delta interactions in immature thymocytes raises the important issue of whether high avidity Notch–Delta binding correlates with Notch signaling. In this respect, analysis of Notch target genes in DN thymocytes stimulated by DL1 or DL4 (expressed on OP9 cells or immobilized on plastic) revealed a considerably better induction of Deltex1 by DL4, consistent with the hierarchy of ligand binding. However, induction of Hes1 in DN thymocytes by DL1 and DL4 was comparable. Collectively, these data favor a scenario in which differences in Notch–Delta binding avidity translate into differences in some Notch signaling outcomes but not in others, presumably because activation of downstream Notch signaling pathways is hierarchal in nature. According to this model, the ability of N1–DL1 interactions to drive T cell lineage commitment and maturation on OP9 cells in vitro would reflect a low-threshold Notch signaling requirement.
Finally, it is worth noting that the hierarchy of Notch–Delta interactions described in this study has potential implications for T cell lineage commitment and maturation under physiological conditions in the thymus. In this context, it is of particular interest that N2 is capable of promoting T cell commitment via interaction with DL1 (but not DL4) both on OP9 stromal cells and during extrathymic T cell development in the spleen. This raises the obvious question of why N2 cannot compensate for N1 during T cell lineage commitment in the thymus. Because N2 is expressed on HSCs and early intrathymic T cell precursors, one possible explanation for the inability of N2 to support T cell commitment would be that DL1 is not present or available in the thymus. Because N2 cannot promote T cell commitment via DL4 even in the sensitive OP9 stromal system, it is unlikely that N2–DL4 interactions would induce T cell development in vivo. This scenario would therefore imply that DL4 is in fact the physiological ligand for N1 during intrathymic T cell development and, more generally, that tissue-specific compartmentalization of Delta family members is a mechanism to assure Notch receptor–ligand specificity in cell fate determination.
A second aspect of hierarchal Notch–Delta interactions that is relevant to the identification of the physiological ligand of N1 during thymus development is the unique ability of DL4 to specifically bind to immature thymocytes with apparent high avidity. Conditional inactivation of N1 in BM precursors (8, 9) and at early stages of intrathymic development (25) has clearly demonstrated that signaling via N1 is required in immature DN thymocytes until they have completed VDJβ rearrangement at the CD25+ DN3 stage, whereas later N1 inactivation (from the DN4 stage onwards) has no impact on subsequent thymus development (36). Interestingly, binding of DL4 by N1 in thymus subsets closely parallels this functional requirement, because DL4 binding is high from the DN1 to DN3 stages and declines in DN4 to become undetectable in subsequent DP and SP stages. This result is again consistent with the hypothesis that DL4 is the physiological N1 ligand responsible for both T cell lineage commitment and subsequent thymic maturation. Moreover, the strict correlation between N1–DL4 binding and N1 function during this process further suggests that N1 signaling on developing thymocytes is regulated at the level of ligand binding. Because N1–DL4 binding activity appears to be relatively independent of Lfng (at least in transfected 293T cells), it could be speculated that N1 function during thymic maturation is largely controlled at the level of N1 expression.
Expression studies of DL1 and DL4 in the thymus, though not definitive, also favor the hypothesis that DL4 may be the physiological N1 ligand for T-lineage commitment and maturation. Thus, semiquantative PCR analysis indicates that DL4 is more strongly expressed than DL1 in the embryonic (18) as well as adult (15, 30) thymus epithelium. More convincingly, in situ hybridization studies, as well as lacZ gene–targeted (knock-in) reporter mice demonstrated clearly that DL4 is expressed at relatively high levels in situ in both embryonic and adult thymus (13, 16, 17), whereas DL1 expression is barely or not detectable (unpublished data) (16, 37). At the protein level, one group has reported broad expression of DL1 in the adult thymus (14); however, the specificity of the polyclonal anti-DL1 antibody used in that study has been challenged (15). Collectively, these data are consistent with the possibility that DL4 rather than DL1 is the physiological thymic ligand for N1. Indeed, conditional inactivation of DL1 in the thymic epithelium does not impair T cell development (30). Nevertheless, it remains possible that DL1 and DL4 function redundantly in N1-mediated T cell lineage commitment. Conditional gene targeting of DL4 will be required to definitively resolve this important issue.
MATERIALS AND METHODS
Mice and induction of the Cre-mediated inativation of the floxed N1 and N2 genes.
Experiments were performed according to Swiss guidelines and authorized by the veterinary authorities of the Canton de Vaud (authorization nos. 1099.2 and 1099.3). CD45.1+ C57BL/6 mice were purchased from the Jackson Laboratory. N1lox/lox&Mx-Cre mice were generated as previously described (8). For N2lox/lox mice, the generation of the N2 targeting vector was based on a genomic DNA fragment including the exons b to h and the 5′ part of exon i (according to the nomenclature previously described [38]) from the mouse N2 locus. Exons d and e (coding for the C-terminal part of the RAM23 domain and nuclear localization sequence) were flanked by loxP sites. Cre recombinase–mediated N2 inactivation results in the expression of a truncated N2 protein, lacking the intracellular part. The generated mice carrying the loxP-targeted exons d and e are referred to here as the floxed N2 allele (N2lox).
Generation of the N2 targeting vector and targeting of embryonic stem (ES) cells.
Construction of the targeting vector based on the genomic N2 phage DNA clone 1NT2-2 (provided by Y. Hamada, National Institute of Basic Biology, Okazaki, Japan) containing the distal exons a to i (exon c turned out to consist of two small neighboring exons, designated by us as c1 and c2). A 9.5-kb SphI fragment from 1NT2-2 encompassing N2 exons b to h and the 5′ part from exon i was subcloned into the backbone vector pHEBOpl-mod, which was previously created from the plasmid pHEBO (39) by replacement of a 630-bp ClaI/SalI fragment with a polylinker sequence and subsequent deletion of a 2.9-kb MluI fragment. Within the genomic N2 SphI fragment, insertions were made at three positions: (a) at the BsrBI site between exons c2 and d, a loxP-flanked neomycin resistance gene cassette (NeoR) from pEasyFlox (provided by Marat Alimzhanov, Harvard Medical School, Boston, MA; unpublished data; reference 40) containing an additional SacI/SstI recognition site between the 5′ loxP site and NeoR was inserted; (b) at the XhoI site between exons e and f, a loxP sequence with a SacI/SstI recognition site at its 3′ end was inserted; and (c) a thymidine-kinase expression cassette (XhoI/PvuI fragment from pEasyFlox) was cloned into the SphI site at the 3′ end of the genomic N2 sequence (within exon i).
The final targeting vector pU1496-21 was sequenced, linearized by NotI, and electroporated into BALB/c-derived ES cells. G418-resistant and ganciclovir-sensitive colonies were screened for homologous recombination by Southern blot analysis. The cellular DNA was digested with SstI (a SacI isoschizomer) and hybridized with a specific N2 probe (a 490-bp SacI/SphI fragment located just upstream of the 5′ end of the genomic N2 sequence in the targeting vector). The WT N2 and the targeted N2 alleles were identified as 7.2- and 2.9-kb fragments, respectively. Clones with homologous recombination were further confirmed by hybridization with an internal probe (NeoR) and a 3′ external probe (271-bp SphI/SacI fragment from exon i). Of 768 analyzed ES cell clones, four exhibited correct recombination with the targeting vector.
In one correctly targeted ES cell clone (1G10), the loxP-flanked NeoR was deleted in vitro by transfection with the cre expression vector pIC-cre (41). The resulting single-cell clones were screened for correct deletion of the NeoR cassette by Southern blotting, as decribed in the previous paragraph. The floxed N2 allele could be identified as a 5.7-kb fragment. Two of the ES cell clones with NeoR deletions were reconfirmed by sequencing and injected into C57BL/6 blastocysts, which were then transferred into foster mothers to obtain chimeric mice. Floxed N2 gene–targeted mice were provided by K. Rajewsky (Harvard Medical School, Boston, MA).
Activation of the Cre recombinase was performed as previously described (8, 9). In brief, Ctrl, (N1lox/lox), N1lox/lox&Mc-Cre, N2lox/lox&Mx-Cre, and N1/N2lox/lox&Mx-Cre mice received five i.p. injections of 250 μg polyI-polyC (pIpC; Sigma-Aldrich) at 2-d intervals. Competitive mixed BM chimeras were set up as previously described (9). In brief, lethally irradiated mice (950 rads 24 h before transfer) that had been treated i.p. 48 h before BM transplantation with 100 μg anti-NK1.1 monoclonal antibodies were reconstituted with a 1:2 mixture (5 × 106:10 × 106) of CD45.1+ WT and Ctrl, N1lox/lox&Mc-Cre, N2lox/lox&Mx-Cre, or N1/N2lox/lox&Mx-Cre BM for mixed chimeras. Mice were maintained on antibiotics (Bactrim) containing water, and reconstitution of BM and lymphoid organs by donor-derived cells was analyzed 8 wk later.
Flow cytometry and cell sorting.
The following monoclonal antibody conjugates were purchased from eBioscience: CD117 (2B8)-PE and -PE-Cy5.5; Sca-1 (D7)-PE and -APC; CD19 (MB-19.1)-PE and (6D5)-PE-Cy5.5; B220 (RA3.6B2)-PE-Cy5.5 and –Alexa Fluor 647; CD44 (IM781)-PE-Cy5.5; CD25 (PC61)-APC; CD4 (L3T4)-PE-Cy5.5; CD45.2 (104)-PE-Cy5.5; TCRβ (H57)-PE and -APC; CD161 (PK136)-FITC; CD90.1 (HIS15)-PE; and CD90.2 (30H12)-PE. TCRγδ (GL3)-PE and TCRβ (H57)-biotin were purchased from BD Biosciences. CD25 (PC61)-PE was purchased from Caltag. CD19 (ID3)–Alexa Fluor 647; B220 (RA3.6B2)–Alexa Fluor 647; CD4 (GK1.5)-FITC and -PE; CD8α (53.6.7)-FITC and –Alexa Fluor 647; CD45.1 (A20)–Alexa Fluor 647; CD45.2 (104)–Alexa Fluor 647; TCRβ (H57)-FITC; TCRγδ (GL3)-FITC; Gr1 (RB6.8C5)-FITC; Ter119-FITC; CD11b (M1/70)-FITC, -PE, and –Alexa Fluor 647; and CD3 (17A2)-FITC were purified from hybridoma supernatants and conjugated in our laboratory according to standard protocols. Alexa Fluor 647 conjugates were prepared using the appropriate Alexa Fluor protein labeling kits (Invitrogen). APC and PE conjugates were prepared using kits purchased from Prozyme. Intracellular staining for TCRβ was performed as previously described (42). Single-cell suspensions were stained with the respective antibodies and analyzed using a FACSCalibur or FACScanto flow cytometer (Becton Dickinson). The cells were sorted with a FACSVantage or a FACSAria flow cytometer (Becton Dickinson). Dead cells and debris were eliminated by appropriate gating on forward and side scatter. The data were analyzed using either CellQuest Pro (BD Biosciences) or FlowJo (TreeStar, Inc.) software.
OP9 cell co-cultures.
OP9 stromal cells engineered to express GFP and the mouse DL1 gene (OP9-DL1 cells, provided by J.C. Zuniga-Pflücker, University of Toronto, Toronto, Canada) or GFP and the mouse DL4 gene (OP9-DL4 cells provided by A. Cumano, Institut Pasteur, Paris, France) were cultured in αMEM supplemented with 20% FBS (Sigma-Aldrich). HSCs were isolated from adult mouse BM and sorted as lin−CD117hiSca-1hi. HSCs were seeded at 4 × 103 cells/well onto 80% confluent monolayers of OP9 cells (24-well plates) in DMEM with 10% FBS. Every third day, 1 ml of the culture supernatant was exchanged with fresh medium. Co-cultures were harvested by pipetting at the time points indicated in the figures, and contaminating OP9 cells were eliminated by filtering the lymphocytes through a 70-μm cell strainer (BD Biosciences) before replating or flow cytometric analysis. All co-cultures were performed in the presence of 5 ng/ml rmIL-7 and rhFlt3L (PeproTech).
Semiquantitative RT-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen), and semiquantitative PCR was performed using the Onestep RT-PCR kit (QIAGEN). All PCR reactions were performed using the same serially diluted RNA samples normalized to an α tubulin– or hypoxanthine guanine phosphoribosyl transferase (HPRT)–specific signal. Gene–specific primer sequences were as follows: N1, (forward) 5′-TGTGACAGCCAGTGCAACTC-3′ and (reverse) 5′-GCAGTGCTTCCAGAGTGCCA-3′; N2, (forward) 5′-ACATCATCACAGACTTGGTC-3′ and (reverse) 5′-GGCAGCTGCTGTCAATAATG-3′; tubulin, (forward) 5′-TCACTGTGCCTGAACTTACC-3′ and (reverse) 5′-GGAACATAGCCGTAAACTGC-3′; mouse and human HPRT, (forward) 5′-AAGGAGATGGGAGGCCATCAC-3′ and (reverse) 5′-CTTGTCTGGAATTTCAAATCCAAC-3′; Deltex1, (forward) 5′-CACTGGCCCTGTCCACCCAGCCTTGGCAGG-3′ and (reverse) 5′-GGGAAGGCGGGCAACTCAGGCCTCAGG-3′; Hes1, (forward) 5′-ATCATGGAGAAGAGGCGAAGGG-3′ and (reverse) 5′-TGATCTGGGTCATGCAGTTGG-3′; and mouse and human Lfng, (forward) 5′-CGCGCCACAAGGAGATGACGTTC-3′ and (reverse) 5′-TGGGCACCTGCTGCAGGTTCT-3′. PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. All PCR products shown correspond to the expected molecular size (Figs. 1, 4, 6, and 7).
Purified DL1- and DL4-IgG were also used to stimulate in vitro DN thymocytes. In brief, 24-well plates were coated with 10 μg/ml protein A overnight at 4°C. 10 μg/ml of purified DL1- and DL4-IgG was added to the plates for 1 h at 4°C. Lineage-depleted DN thymocytes were subsequently plated on DL fusion protein–coated plates at 2 × 106 cells/well. After 20 h of culture, DN thymocytes were harvested to analyze gene expression by semiquantitative RT-PCR.
Spleen CFU (CFU-S) assay.
CD45.1+ C57BL/6 mice were exposed to lethal whole-body irradiation (950 rads) from a 137Cs source and maintained on water containing antibiotics (Bactrim). The next day, total BM cells (12 × 104) from either Ctrl, N1−/−, N2−/−, or N1N2−/− mice were injected i.v. via the retroorbital sinus. After 12 d, spleens of the recipient mice were removed, and single-cell suspensions were prepared for FACS analysis.
In vitro stimulation of 5-FU–treated BM cells.
1 wk after the last polyI-polyC injection, N1lox/lox and N1lox/lox&Mx-Cre mice were treated with 3 mg/20 g body weight of 5-fluorouracil (Sigma-Aldrich). 5 d later, BM cells were harvested and, after red blood cell lysis, single-cell suspensions were prepared in stem cell–activating (SA) medium containing IMDM supplemented with 10% FBS, 100 ng/ml rmSCF (R&D Systems), 50 ng/ml rmTPO (R&D Systems), and 50 ng/ml rmFlt-3L (R&D Systems). The cells were incubated overnight at 37°C in 5% CO2 before the retroviral infection.
Retrovirus production and infection procedure.
Empty (MIG) or recombinant (DL1 and DL4) retroviruses (provided by A. Freitas, Institut Pasteur, Paris, France) were obtained after transfection of Bosc23 packaging cells using lipofectamine 2000 (Invitrogen). Retrovirus-containing supernatants were collected 48 h after transfection. 3-cm petri dishes were coated with 1 ml of RetroNectin (r-fibronectin fragment CH-296; TaKara) solution (12.5 μg/ml in PBS) for 2 h at room temperature. After removal of the RetroNectin, 2 ml PBS + 2% BSA was added to each dish for 30 min. After washing the plates with PBS, the retroviral supernatant was added to the coated plates for 1 h at 37°C. The prestimulated BM cells were spun down, resuspended in 1 ml of fresh SA medium, and added to the retroviral supernatant. The next day, 1–3 × 106 cells were injected i.v. into lethally irradiated CD45.1+ C57BL/6 mice.
To ensure similar functionality and/or expression of DL1 and DL4, the retroviruses were assessed before BM infection by Western blot analysis, as well as by infecting OP9 cells that were subsequently tested for their ability to induce T cell development in vitro. Based on these criteria, DL1 and DL4 retroviruses were equivalent (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20061442/DC1).
Expression plasmids and transfections.
The mouse cDNA coding for the extracellular domain of either DL1 or DL4 was cloned via HindIII/BamHI and EcoRI/SalI, respectively, into the PS 521 expression vector (43) to generate DL1- and DL4-IgG fusion proteins. The corresponding expression vectors were transfected into 293T cells using the calcium-phosphate method, and IgG fusion proteins were subsequently purified over protein A columns according to the manufacturer's instructions (HiTrap rProtein A FF; GE Healthcare). Purity of the fusion proteins was verified by Coomassie blue staining and Western blot analysis. Full-length N1 and N2 cDNAs were cloned in frame into the pEGFP-N1 expression vector (CLONTECH Laboratories, Inc.). The mouse cDNA coding for the Lfng was cloned via BamHI into pcDNA3.1− (Invitrogen). 293T cells were transiently transfected with 5 μg N1- and N2-EGFP expression vectors, respectively, with or without the Lfng expression vector. 48 h later, the cells were indirectly stained with DL1- or DL4-IgG fusion proteins (0.5 μg/106 cells). Binding of the fusion proteins to the Notch receptors was detected using biotinylated anti–human IgG antibodies.
Online supplemental material.
Fig. S1 shows a functional test for DL1- and DL4-expressing retroviruses. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20061442/DC1.
Supplemental Material
Acknowledgments
We thank F. Grosjean and E. Devevre for cell sorting, P. Schneider for the PS521 expression vector and helpful suggestions, and A. Wilson for discussions and critical reading of the manuscript.
This work was supported in part by the Swiss National Science Foundation (Förderungsprofessur to F. Radtke), a Marie Heim-Vögtlein Fellowship (PMP DB-110307/1 to E. Fiorini), the Swiss Cancer League, and grants from the Deutsche Forschungsgemeinschaft (SFP 243, STR-461/3-2, und SFB 684).
The authors have no conflicting financial interests.
Abbreviations used: DL1, Delta1; DN, double negative; DP, double positive; EGFP, enhanced GFP; ES, embryonic stem; HPRT, hypoxanthine guanine phosphoribosyl transferase; HSC, hematopoietic stem cell; ISP, immature SP; KLS, CD117+lin−Sca1+; Lfng, Lunatic fringe; MZB cell, marginal zone B cell; N1, Notch1; SP, single positive.
V. Besseyrias, E. Fiorini, L.J. Strobl, and U. Zimber-Strobl contributed equally to this work.
References
- 1.Rothenberg, E.V. 2000. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 10:370–379. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey, D.I., and A. Zlotnik. 1993. Control points in early T-cell development. Immunol. Today. 14:547–553. [DOI] [PubMed] [Google Scholar]
- 3.Rodewald, H.R., and H.J. Fehling. 1998. Molecular and cellular events in early thymocyte development. Adv. Immunol. 69:1–112. [DOI] [PubMed] [Google Scholar]
- 4.Radtke, F., A. Wilson, S.J. Mancini, and H.R. MacDonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247–253. [DOI] [PubMed] [Google Scholar]
- 5.Maillard, I., T. Fang, and W.S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945–974. [DOI] [PubMed] [Google Scholar]
- 6.Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- 7.Haines, N., and K. Irvine. 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4:786–797. [DOI] [PubMed] [Google Scholar]
- 8.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 9.Wilson, A., H.R. MacDonald, and F. Radtke. 2001. Notch1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, H., K. Tanigaki, N. Yamamoto, K. Kuroda, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. [DOI] [PubMed] [Google Scholar]
- 11.Hasserjian, R.P., J.C. Aster, F. Davi, D.S. Weinberg, and J. Sklar. 1996. Modulated expression of notch1 during thymocyte development. Blood. 88:970–976. [PubMed] [Google Scholar]
- 12.Felli, M.P., M. Maroder, T.A. Mitsiadis, A.F. Campese, D. Bellavia, A. Vacca, R.S. Mann, L. Frati, U. Lendahl, A. Gulino, and I. Screpanti. 1999. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 11:1017–1025. [DOI] [PubMed] [Google Scholar]
- 13.Yan, X.Q., U. Sarmiento, Y. Sun, G. Huang, J. Guo, T. Juan, G. Van, M.Y. Qi, S. Scully, G. Senaldi, and F.A. Fletcher. 2001. A novel Notch ligand, Dll4, induces T-cell leukemia/lymphoma when overexpressed in mice by retroviral-mediated gene transfer. Blood. 98:3793–3799. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt, T.M., M. Ciofani, H.T. Petrie, and J.C. Zuniga-Pflucker. 2004. Maintenance of T cell specification and differentiation requires recurrent Notch receptor–ligand interactions. J. Exp. Med. 200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehar, S.M., J. Dooley, A.G. Farr, and M.J. Bevan. 2005. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 105:1440–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukamoto, N., M. Itoi, M. Nishikawa, and T. Amagai. 2005. Lack of Delta like 1 and 4 expressions in nude thymus anlages. Cell. Immunol. 234:77–80. [DOI] [PubMed] [Google Scholar]
- 17.Benedito, R., and A. Duarte. 2005. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr. Patterns. 5:750–755. [DOI] [PubMed] [Google Scholar]
- 18.Mohtashami, M., and J.C. Zuniga-Pflucker. 2006. Three-dimensional architecture of the thymus is required to maintain delta-like expression necessary for inducing T cell development. J. Immunol. 176:730–734. [DOI] [PubMed] [Google Scholar]
- 19.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, K. Kumano, et al. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 18:675–685. [DOI] [PubMed] [Google Scholar]
- 20.Krebs, L.T., Y. Xue, C.R. Norton, J.P. Sundberg, P. Beatus, U. Lendahl, A. Joutel, and T. Gridley. 2003. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis. 37:139–143. [DOI] [PubMed] [Google Scholar]
- 21.Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnell, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, and W.S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. [DOI] [PubMed] [Google Scholar]
- 22.De Smedt, M., K. Reynvoet, T. Kerre, T. Taghon, B. Verhasselt, B. Vandekerckhove, G. Leclercq, and J. Plum. 2002. Active form of Notch imposes T cell fate in human progenitor cells. J. Immunol. 169:3021–3029. [DOI] [PubMed] [Google Scholar]
- 23.Dorsch, M., G. Zheng, D. Yowe, P. Rao, Y. Wang, Q. Shen, C. Murphy, X. Xiong, Q. Shi, J.C. Gutierrez-Ramos, et al. 2002. Ectopic expression of Delta4 impairs hematopoietic development and leads to lymphoproliferative disease. Blood. 100:2046–2055. [PubMed] [Google Scholar]
- 24.de La Coste, A., E. Six, N. Fazilleau, L. Mascarell, N. Legrand, M.P. Mailhe, A. Cumano, Y. Laabi, and A.A. Freitas. 2005. In vivo and in absence of a thymus, the enforced expression of the notch ligands delta-1 or delta-4 promotes T cell development with specific unique effects. J. Immunol. 174:2730–2737. [DOI] [PubMed] [Google Scholar]
- 25.Wolfer, A., A. Wilson, M. Nemir, H.R. MacDonald, and F. Radtke. 2002. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity. 16:869–879. [DOI] [PubMed] [Google Scholar]
- 26.Mancini, S.J., N. Mantei, A. Dumortier, U. Suter, H.R. Macdonald, and F. Radtke. 2005. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 105:2340–2342. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, R., Y. Lan, H.D. Chapman, C. Shawber, C.R. Norton, D.V. Serreze, G. Weinmaster, and T. Gridley. 1998. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12:1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaleco, A.C., H. Neves, E. Hooijberg, P. Gameiro, N. Clode, M. Haury, D. Henrique, and L. Parreira. 2001. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J. Exp. Med. 194:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt, T.M., and J.C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 30.Hozumi, K., N. Negishi, D. Suzuki, N. Abe, Y. Sotomaru, N. Tamaoki, C. Mailhos, D. Ish-Horowicz, S. Habu, and M.J. Owen. 2004. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat. Immunol. 5:638–644. [DOI] [PubMed] [Google Scholar]
- 31.Maillard, I., B.A. Schwarz, A. Sambandam, T. Fang, O. Shestova, L. Xu, A. Bhandoola, and W.S. Pear. 2006. Notch-dependent T-lineage commitment occurs at extrathymic sites following bone marrow transplantation. Blood. 107:3511–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancrin, C., E. Schneider, F. Lambolez, M.L. Arcangeli, C. Garcia-Cordier, B. Rocha, and S. Ezine. 2002. Major T cell progenitor activity in bone marrow–derived spleen colonies. J. Exp. Med. 195:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcangeli, M.L., C. Lancrin, F. Lambolez, C. Cordier, E. Schneider, B. Rocha, and S. Ezine. 2005. Extrathymic hemopoietic progenitors committed to T cell differentiation in the adult mouse. J. Immunol. 174:1980–1988. [DOI] [PubMed] [Google Scholar]
- 34.Visan, I., J.B. Tan, J.S. Yuan, J.A. Harper, U. Koch, and C.J. Guidos. 2006. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nat. Immunol. 7:634–643. [DOI] [PubMed] [Google Scholar]
- 35.Till, J.E., and C.E. McCulloch. 1961. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14:213–222. [PubMed] [Google Scholar]
- 36.Wolfer, A., T. Bakker, A. Wilson, M. Nicolas, V. Ioannidis, D.R. Littman, P.P. Lee, C.B. Wilson, W. Held, H.R. MacDonald, and F. Radtke. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2:235–241. [DOI] [PubMed] [Google Scholar]
- 37.Visan, I., J.S. Yuan, J.B. Tan, K. Cretegny, and C.J. Guidos. 2006. Regulation of intrathymic T-cell development by Lunatic Fringe-Notch1 interactions. Immunol. Rev. 209:76–94. [DOI] [PubMed] [Google Scholar]
- 38.Hamada, Y., Y. Kadokawa, M. Okabe, M. Ikawa, J.R. Coleman, and Y. Tsujimoto. 1999. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 126:3415–3424. [DOI] [PubMed] [Google Scholar]
- 39.Sugden, B., K. Marsh, and J. Yates. 1985. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol. Cell. Biol. 5:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenten, D., V.L. Gerlach, C. Guo, S. Velasco-Miguel, C.L. Hladik, C.L. White, E.C. Friedberg, K. Rajewsky, and G. Esposito. 2002. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur. J. Immunol. 32:3152–3160. [DOI] [PubMed] [Google Scholar]
- 41.Gu, H., Y.R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 73:1155–1164. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, A., M. Capone, and H.R. MacDonald. 1999. Unexpectedly late expression of intracellular CD3epsilon and TCR gammadelta proteins during adult thymus development. Int. Immunol. 11:1641–1650. [DOI] [PubMed] [Google Scholar]
- 43.Schneider, P. 2000. Production of recombinant TRAIL and TRAIL receptor: Fc chimeric proteins. Methods Enzymol. 322:325–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.