Abstract
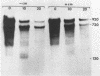
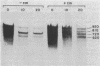
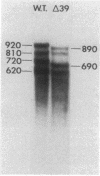
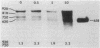
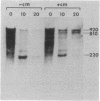
cat-86 is a promoter-deficient plasmid gene that encodes chloramphenicol acetyltransferase (CAT). Insertion of a promoter at a site 144 base pairs 5' to the cat-86 coding sequence activates transcription of the gene and allows cat-86 to specify chloramphenicol-inducible CAT activity in Bacillus subtilis. Induction of cat-86 by chloramphenicol has been shown to result from a regulatory event that activates translation of cat-86 mRNA that is present in cells before the addition of inducer (E. J. Duvall and P. S. Lovett, Proc. Natl. Acad. Sci. USA 83:3939-3943, 1986). In the present study we show an unusual property of cat-86 mRNA. Full-length cat-86 transcripts, consisting of 920 nucleotides (nt), are cleaved in B. subtilis to yield two predominant fragmentation products: an 810-nt species that lacks sequences present at the 5' end of the 920-nt species and a 720-nt species that lacks sequences present at the 3' end of the 920-nt species. A third fragmentation product consisting of 620 nt may result from the cleavage of a single 920-nt transcript at both the 5' and 3' ends. The sequences which are missing from the 720- and 620-nt species suggest that these transcripts cannot be translated into functional CAT. The 810-nt species lacks sequences from the 5' regulatory region, and it is not yet certain whether or not translation of this species can be induced by chloramphenicol. The ratio of 920-nt molecules/720-nt molecules in rifampin-treated cells is increased when the cells are grown in chloramphenicol. Therefore, induction may partially stabilize full-length cat-86 transcripts against inactivation by a novel processing-like system.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. Analysis of the regulatory sequences needed for induction of the chloramphenicol acetyltransferase gene cat-86 by chloramphenicol and amicetin. J Bacteriol. 1986 Sep;167(3):842–849. doi: 10.1128/jb.167.3.842-849.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Kaufman J. D., Lovett P. S. Chloramphenicol-induced translation of cat-86 mRNA requires two cis-acting regulatory regions. J Bacteriol. 1985 Nov;164(2):696–703. doi: 10.1128/jb.164.2.696-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Lovett P. S. A transcription termination signal immediately precedes the coding sequence for the chloramphenicol-inducible plasmid gene cat-86. Mol Gen Genet. 1985;199(1):70–75. doi: 10.1007/BF00327512. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Blundell M., Kennell D. Evidence for endonucleolytic attack in decay of lac messenger RNA in Escherichia coli. J Mol Biol. 1974 Feb 25;83(2):143–161. doi: 10.1016/0022-2836(74)90385-4. [DOI] [PubMed] [Google Scholar]
- Brückner R., Matzura H. Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 1985 Sep;4(9):2295–2300. doi: 10.1002/j.1460-2075.1985.tb03929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon W. H., Weisblum B. Post-transcriptional regulation of chloramphenicol acetyl transferase. J Bacteriol. 1984 May;158(2):543–550. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. E. coli RNases: making sense of alphabet soup. Cell. 1985 Apr;40(4):731–732. doi: 10.1016/0092-8674(85)90330-7. [DOI] [PubMed] [Google Scholar]
- Downard J. S., Whiteley H. R. Early RNAs in SP82- and SP01-infected Bacillus subtilis may be processed. J Virol. 1981 Mar;37(3):1075–1078. doi: 10.1128/jvi.37.3.1075-1078.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Lovett P. S. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3939–3943. doi: 10.1073/pnas.83.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y., Imamoto F. Evidence for endonucleolytic cleavage at the 5'-proximal segment of the trp messenger RNA in Escherichia coli. Mol Gen Genet. 1979 Apr 17;172(1):25–30. doi: 10.1007/BF00276211. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Evidence for random endonucleolytic cleavages between messages in decay of Escherichia coli trp mRNA. J Mol Biol. 1980 Aug 5;141(2):227–233. doi: 10.1016/0022-2836(80)90388-5. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Models for decay of Escherichia coli lac messenger RNA and evidence for inactivating cleavages between its messages. J Mol Biol. 1979 Dec 5;135(2):369–390. doi: 10.1016/0022-2836(79)90442-x. [DOI] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Duvall E. J., Lovett P. S. Transcription termination signal for the cat-86 indicator gene in a Bacillus subtilis promoter-cloning plasmid. Gene. 1985;37(1-3):83–90. doi: 10.1016/0378-1119(85)90260-4. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Whiteley H. R. Bacillus subtilis RNAase III cleavage sites in phage SP82 early mRNA. Cell. 1983 Jul;33(3):907–913. doi: 10.1016/0092-8674(83)90033-8. [DOI] [PubMed] [Google Scholar]
- Panganiban A. T., Whiteley H. R. Purification and properties of a new bacillus subtilis RNA processing enzyme. Cleavage of phage SP82 mRNA and Bacillus subtilis precursor rRNA. J Biol Chem. 1983 Oct 25;258(20):12487–12493. [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the -35 to -10 spacing. Gene. 1982 Dec;20(2):231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- Schlessinger D., Jacobs K. A., Gupta R. S., Kano Y., Imamoto F. Decay of individual Escherichia coli trp messenger RNA molecules is sequentially ordered. J Mol Biol. 1977 Mar 5;110(3):421–439. doi: 10.1016/s0022-2836(77)80107-1. [DOI] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]