Abstract
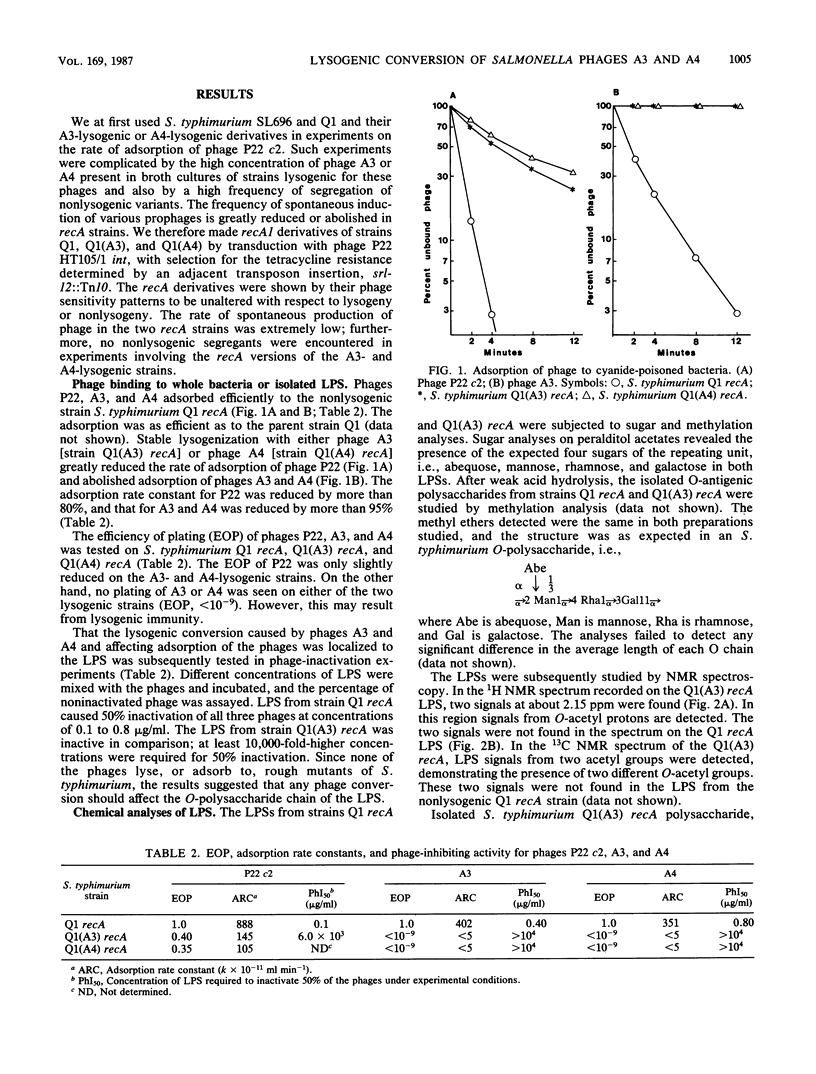
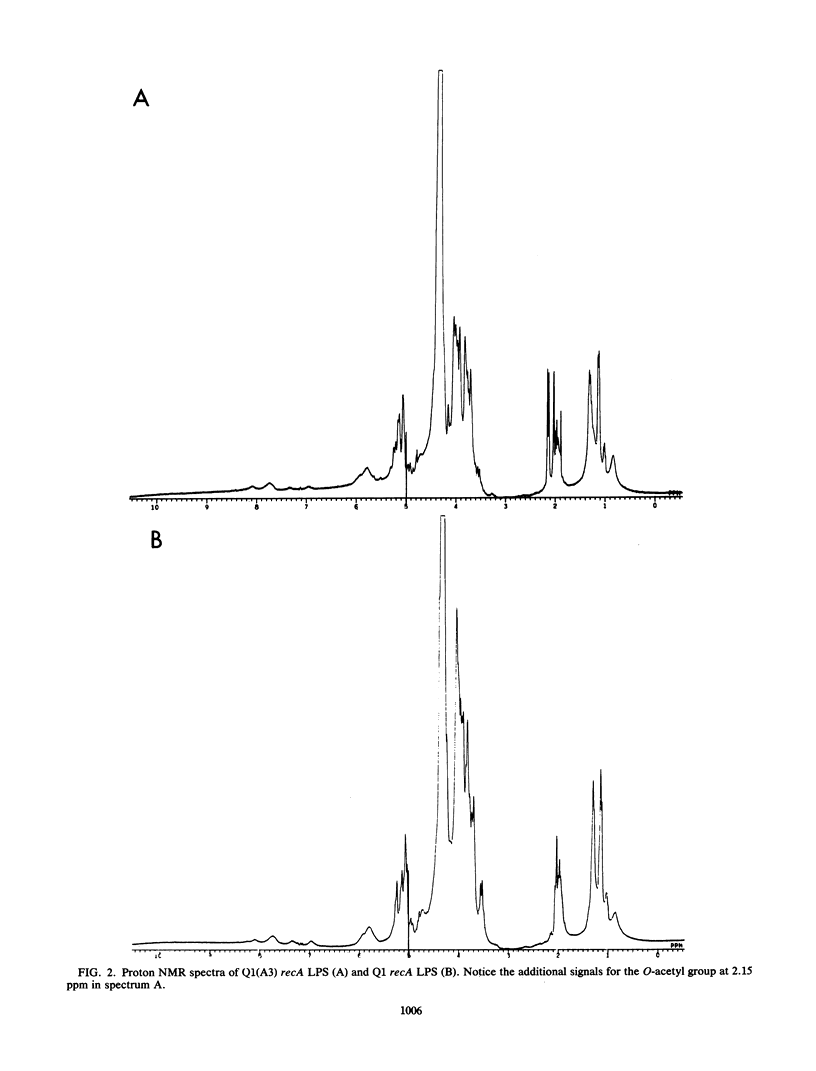
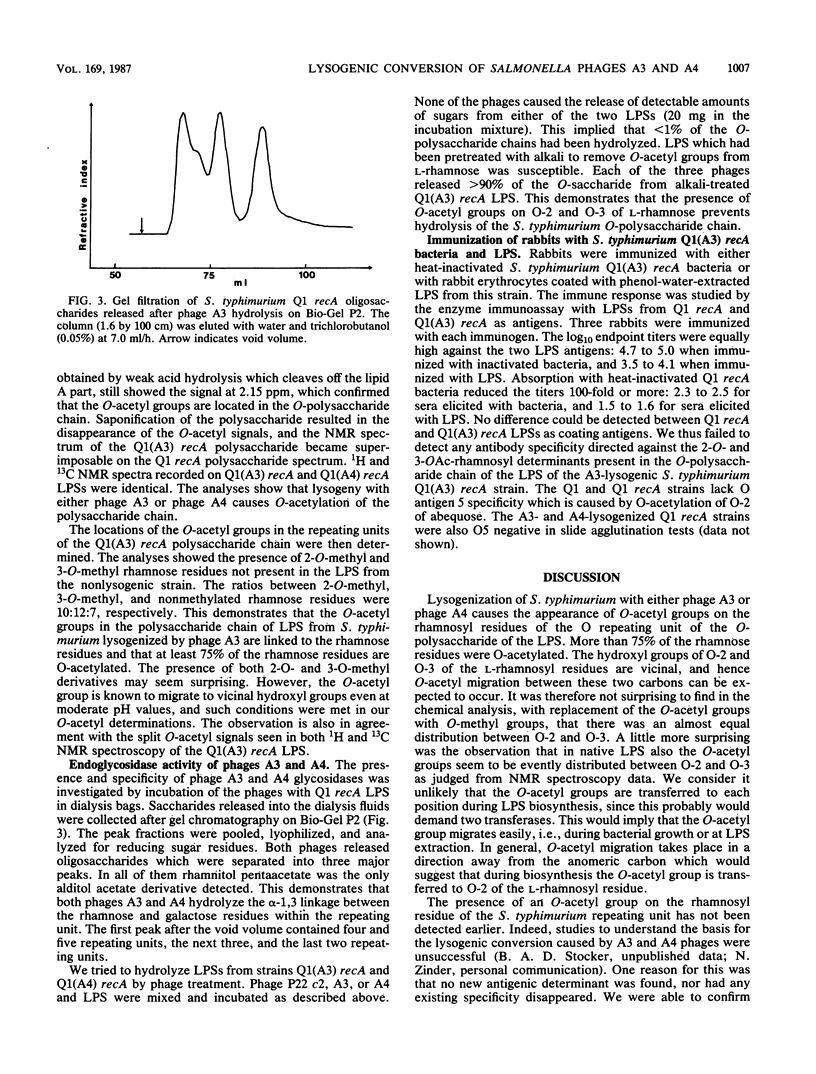
Lysogenization of Salmonella typhimurium with either of the bacteriophages A3 and A4 results in O-acetylation of the L-rhamnose residues of the O-polysaccharide chain of the lipopolysaccharide of the bacterial cell envelope. The O-acetyl group is found on both O-2 and O-3 of the L-rhamnosyl residues. This lysogenic conversion prevents the adsorption of the A3 and A4 phages and also greatly reduces the rate of adsorption of phage P22 to the O-polysaccharide chain as measured by binding studies with whole bacteria. Isolated lipopolysaccharide from A3- and A4-lysogenized bacteria was also inefficient in inactivating these phages: the concentration required for 50% inactivation was 10,000-fold higher than that for lipopolysaccharide from S. typhimurium not lysogenized by any A phage. Binding of phages A3 and A4 is accompanied by hydrolysis of the alpha-1,3 linkage between rhamnose and galactose in the tetrasaccharide repeating unit of the O-polysaccharide. Phage hydrolysis generates saccharides of various lengths, the majority being dodecasaccharides, i.e., equivalent to three repeating units. It is surmised that O-acetylation of the rhamnosyl residue interferes with phage A3, A4, and P22 infection by preventing binding to and hydrolysis of the O-polysaccharide chain, the initial step in the phage infection cycle. The new O-acetyl-rhamnose entities did not elicit specific antibodies in rabbits in accordance with earlier experiences. The O-acetylation of O-2 and O-3 of rhamnose is a new, hitherto unknown, modification of the O-polysaccharide chain of S. typhimurium.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOYD J. S. K. The symbiotic bacteriophages of Salmonella typhi-murium. J Pathol Bacteriol. 1950 Oct;62(4):501–517. doi: 10.1002/path.1700620402. [DOI] [PubMed] [Google Scholar]
- BOYD J. S., BIDWELL D. E. The type A phages of Salmonella typhimurium: identification by a standardized cross-immunity test. J Gen Microbiol. 1957 Feb;16(1):217–228. doi: 10.1099/00221287-16-1-217. [DOI] [PubMed] [Google Scholar]
- BOYD J. S. Bacteriophage and heredity. Nature. 1954 May 29;173(4413):1050–1051. doi: 10.1038/1731050a0. [DOI] [PubMed] [Google Scholar]
- BRADLEY P. L., BOYD J. S. K. Readsorption of phage produced in cultures of lysogenic strains of Salmonella typhi-murium. J Pathol Bacteriol. 1952 Oct;64(4):891–892. doi: 10.1002/path.1700640421. [DOI] [PubMed] [Google Scholar]
- Bock K., Meldal M., Bundle D. R., Iversen T., Garegg P. J., Norberg T., Lindberg A. A., Svenson S. B. The conformation of Salmonella O-antigenic polysaccharide chains of serogroups A, B, and D1 predicted by semi-empirical, Hard-Sphere (HSEA) calculations. Carbohydr Res. 1984 Jul 15;130:23–34. doi: 10.1016/0008-6215(84)85267-2. [DOI] [PubMed] [Google Scholar]
- Carlsson H. E., Lindberg A. A., Hammarström S. Titration of antibodies to salmonella O antigens by enzyme-linked immunosorbent assay. Infect Immun. 1972 Nov;6(5):703–708. doi: 10.1128/iai.6.5.703-708.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Evaluation of some extraction methods for the preparation of bacterial lipopolysaccharides for structural analysis. Acta Pathol Microbiol Scand B Microbiol Immunol. 1972;80(5):751–759. doi: 10.1111/j.1699-0463.1972.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Lindberg A. A., Holme T. Influence of O side chains on the attachment of the Felix O-1 bacteriophage to Salmonella bacteria. J Bacteriol. 1969 Aug;99(2):513–519. doi: 10.1128/jb.99.2.513-519.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg A. A., Wollin R., Gemski P., Wohlhieter J. A. Interaction between bacteriophage Sf6 and Shigella flexner. J Virol. 1978 Jul;27(1):38–44. doi: 10.1128/jvi.27.1.38-44.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüderitz O., Staub A. M., Westphal O. Immunochemistry of O and R antigens of Salmonella and related Enterobacteriaceae. Bacteriol Rev. 1966 Mar;30(1):192–255. doi: 10.1128/br.30.1.192-255.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson S. B., Lönngren J., Carlin N., Lindberg A. A. Salmonella bacteriophage glycanases: endorhamnosidases of Salmonella typhimurium bacteriophages. J Virol. 1979 Nov;32(2):583–592. doi: 10.1128/jvi.32.2.583-592.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson R. G., Gemski P., Jr, Stocker B. A. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J Gen Microbiol. 1972 May;70(3):527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- Wollin R., Eriksson U., Lindberg A. A. Salmonella bacteriophage glycanases: endorhamnosidase activity of bacteriophages P27, 9NA, and KB1. J Virol. 1981 Jun;38(3):1025–1033. doi: 10.1128/jvi.38.3.1025-1033.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]



