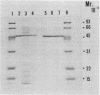
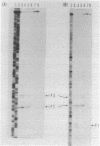
Abstract
We have isolated and sequenced a gene (amy) coding for alpha-amylase (EC 3.2.1.1.) from the Streptomyces hygroscopicus genome (H. Hidaka, Y. Koaze, K. Yoshida, T. Niwa, T. Shomura, and T. Niida, Die Stärke 26:413-416, 1974). Amylase was purified to obtain amino acid sequence information which was used to synthesize oligonucleotide probes. amy-containing Escherichia coli cosmids identified by hybridization did not express amylase activity. Subcloning experiments indicated that amy could be expressed from the lac promoter in E. coli or from its own promoter in S. lividans. The amy nucleotide sequence indicated that it coded for a protein of 52 kilodaltons (478 amino acids). Secreted alpha-amylase contained amino- and carboxy-terminal as well as internal amino acid sequences which were consistent with the nucleotide sequence. The 30-residue leader sequence showed similarities to those found in other procaryotes. The DNA sequence 5' to the amy structural gene contained a sequence complementary to the 3'-terminal sequence of 16S rRNA of S. lividans (M. J. Bibb and S. N. Cohen, Mol. Gen. Genet. 187:265-277, 1982). The transcriptional start points of amy were determined by mung bean nuclease mapping, but the promoter of amy was not similar to the consensus sequence found in other procaryotes.
Full text
PDF
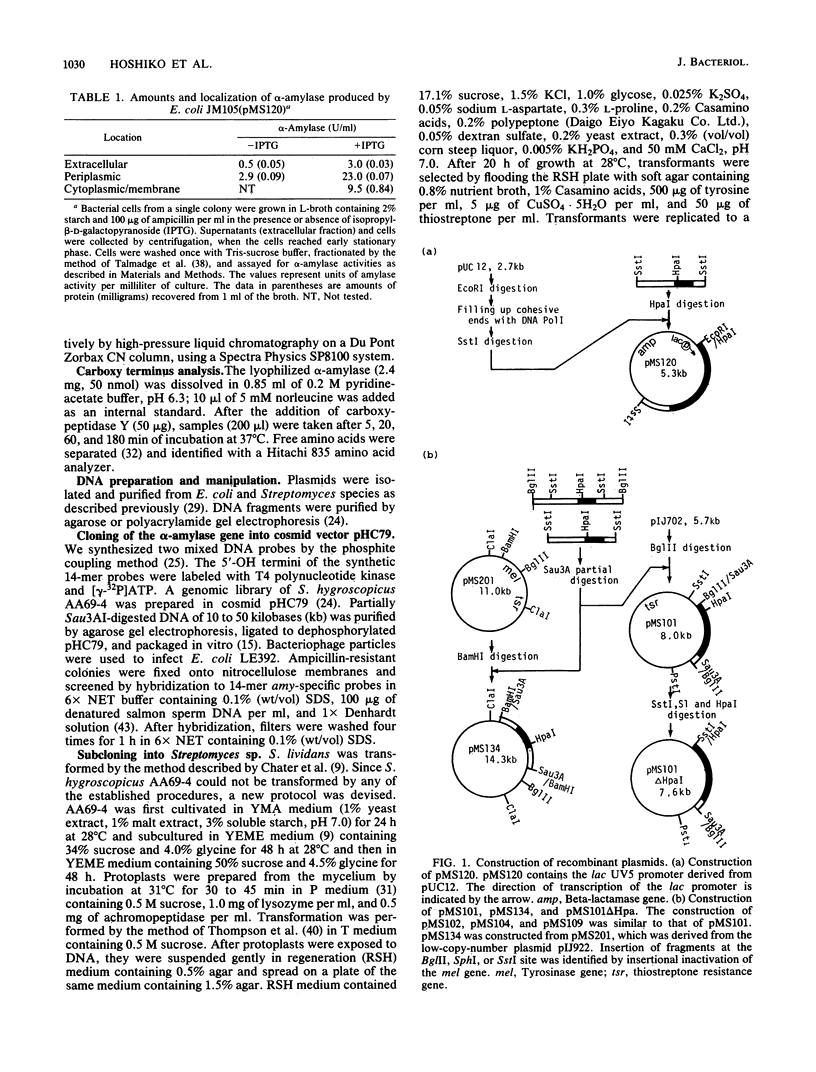

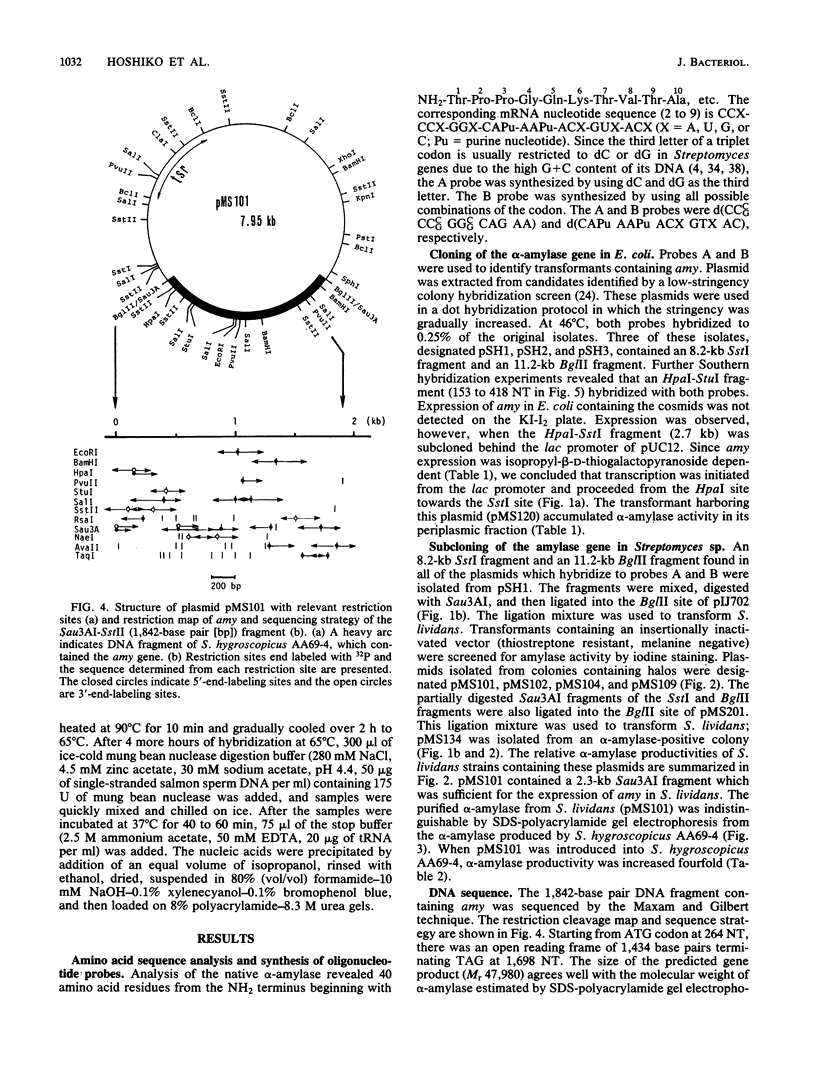




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambartsumyan N. S., Mazo A. M. Elimination of the secondary structure effect in gell sequencing of nucleic acids. FEBS Lett. 1980 Jun 2;114(2):265–268. doi: 10.1016/0014-5793(80)81130-6. [DOI] [PubMed] [Google Scholar]
- Bernan V., Filpula D., Herber W., Bibb M., Katz E. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene. 1985;37(1-3):101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Buttner M. J., Brown N. L. RNA polymerase-DNA interactions in Streptomyces. In vitro studies of a S. lividans plasmid promoter with S. coelicolor RNA polymerase. J Mol Biol. 1985 Sep 5;185(1):177–188. doi: 10.1016/0022-2836(85)90189-5. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T., Thompson C. J. Gene cloning in Streptomyces. Curr Top Microbiol Immunol. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Enquist L. W., Madden M. J., Schiop-Stanley P., Vande Woude G. F. Cloning of herpes simplex type 1 DNA fragments in a bacteriophage lambda vector. Science. 1979 Feb 9;203(4380):541–544. doi: 10.1126/science.216076. [DOI] [PubMed] [Google Scholar]
- Green M. R., Roeder R. G. Definition of a novel promoter for the major adenovirus-associated virus mRNA. Cell. 1980 Nov;22(1 Pt 1):231–242. doi: 10.1016/0092-8674(80)90171-3. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Ihara H., Sasaki T., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a thermophilic alpha-amylase gene: homology between prokaryotic and eukaryotic alpha-amylases at the active sites. J Biochem. 1985 Jul;98(1):95–103. doi: 10.1093/oxfordjournals.jbchem.a135279. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kendall K., Cullum J. Cloning and expression of an extracellular-agarase from Streptomyces coelicolor A3(2) in Streptomyces lividans 66. Gene. 1984 Sep;29(3):315–321. doi: 10.1016/0378-1119(84)90060-x. [DOI] [PubMed] [Google Scholar]
- Kirby K. S., Fox-Carter E., Guest M. Isolation of deoxyribonucleic acid and ribosomal ribonucleic acid from bacteria. Biochem J. 1967 Jul;104(1):258–262. doi: 10.1042/bj1040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydiate D. J., Malpartida F., Hopwood D. A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35(3):223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Nojiri C., Toyama H., Hayashi E., Yamada Y., Nagaoka K. Pock forming plasmids from antibiotic-producing Streptomyces. J Antibiot (Tokyo) 1983 Apr;36(4):429–434. doi: 10.7164/antibiotics.36.429. [DOI] [PubMed] [Google Scholar]
- Nakajima R., Imanaka T., Aiba S. Nucleotide sequence of the Bacillus stearothermophilus alpha-amylase gene. J Bacteriol. 1985 Jul;163(1):401–406. doi: 10.1128/jb.163.1.401-406.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okanishi M., Suzuki K., Umezawa H. Formation and reversion of Streptomycete protoplasts: cultural condition and morphological study. J Gen Microbiol. 1974 Feb;80(2):389–400. doi: 10.1099/00221287-80-2-389. [DOI] [PubMed] [Google Scholar]
- Rinderknecht E., O'Connor B. H., Rodriguez H. Natural human interferon-gamma. Complete amino acid sequence and determination of sites of glycosylation. J Biol Chem. 1984 Jun 10;259(11):6790–6797. [PubMed] [Google Scholar]
- Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1984 Jun 25;259(12):7577–7583. [PubMed] [Google Scholar]
- Robbins P. W., Wirth D. F., Hering C. Expression of the Streptomyces enzyme endoglycosidase H in Escherichia coli. J Biol Chem. 1981 Oct 25;256(20):10640–10644. [PubMed] [Google Scholar]
- Rodgers W. H., Springer W., Young F. E. Cloning and expression of a Streptomyces fradiae neomycin resistance gene in Escherichia coli. Gene. 1982 May;18(2):133–141. doi: 10.1016/0378-1119(82)90111-1. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Talmadge K., Stahl S., Gilbert W. Eukaryotic signal sequence transports insulin antigen in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3369–3373. doi: 10.1073/pnas.77.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Ward J. M., Hopwood D. A. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J Bacteriol. 1982 Aug;151(2):668–677. doi: 10.1128/jb.151.2.668-677.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Williams S. T., Goodfellow M., Alderson G., Wellington E. M., Sneath P. H., Sackin M. J. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983 Jun;129(6):1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]