Abstract
The D-serine deaminase structural (dsdA) and regulatory (dsdC) genes are transcribed with opposite polarity from an intergenic region comprising more than 600 base pairs. The order of genes in the dsd region is supN-dsdA-dsdC-aroC---his. The DNA sequence of the intergenic region has been slightly revised from a previously published version (E. McFall and L. Runkel, J. Bacteriol. 154:1508-1512, 1983). The dsdA gene is preceded by a long open reading frame. The dsdA in vivo transcription start sites for the wild type (base pair +1) and for three phenotypically distinct promoter constitutive mutants were determined by the S1 nuclease method. They are identical and are located about 81 base pairs upstream of the translation start site. D-Serine deaminase regulation is normal in rho mutants. Possible mechanisms for dsdA activation are discussed.
Full text
PDF
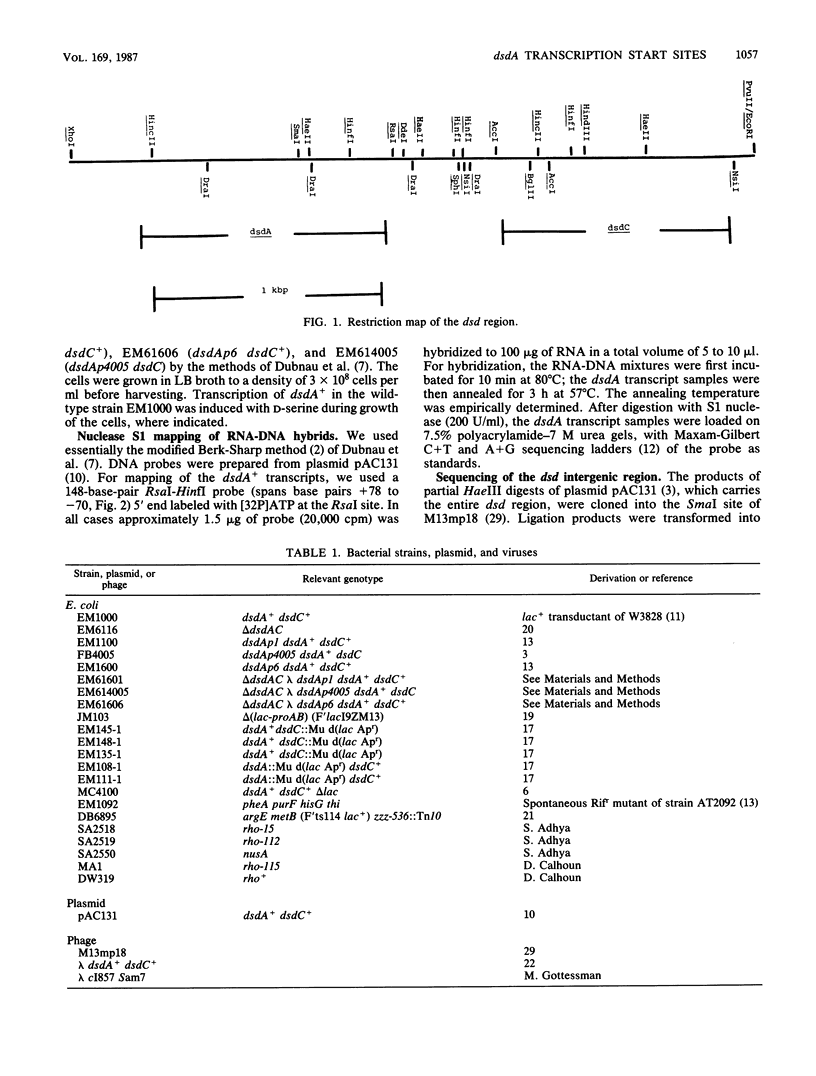

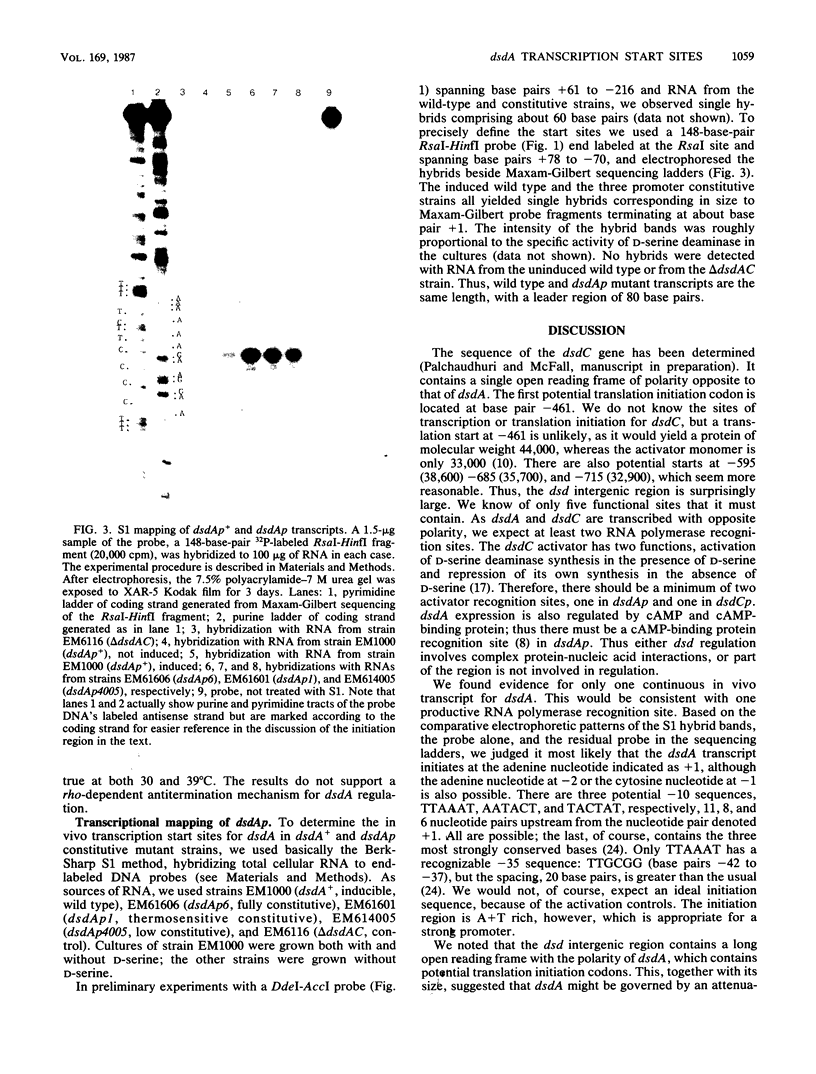

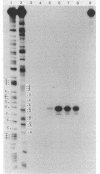
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bloom F. R. Isolation and characterization of catabolite-resistant mutants in the D-serine deaminase system of Escherichia coli K-12. J Bacteriol. 1975 Mar;121(3):1085–1091. doi: 10.1128/jb.121.3.1085-1091.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., McFall E. Escape synthesis of D-serine deaminase in lambda dsdC integration lysogens. J Bacteriol. 1976 Aug;127(2):1028–1029. doi: 10.1128/jb.127.2.1028-1029.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., McFall E., Palchaudhuri S. Physical mapping of the Escherichia coli D-serine deaminase region: contiguity of the dsd structural and regulatory genes. J Bacteriol. 1980 Apr;142(1):174–184. doi: 10.1128/jb.142.1.174-184.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau E., Ramakrishna N., Cabane K., Smith I. Cloning of an early sporulation gene in Bacillus subtilis. J Bacteriol. 1981 Aug;147(2):622–632. doi: 10.1128/jb.147.2.622-632.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Friedman D. I., Olson E. R. Evidence that a nucleotide sequence, "boxA," is involved in the action of the NusA protein. Cell. 1983 Aug;34(1):143–149. doi: 10.1016/0092-8674(83)90144-7. [DOI] [PubMed] [Google Scholar]
- Heincz M. C., Bornstein S. M., McFall E. Purification and characterization of D-serine deaminase activator protein. J Bacteriol. 1984 Oct;160(1):42–49. doi: 10.1128/jb.160.1.42-49.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heincz M. C., McFall E. Role of the dsdC activator in regulation of D-serine deaminase synthesis. J Bacteriol. 1978 Oct;136(1):96–103. doi: 10.1128/jb.136.1.96-103.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E. Escherichia coli K-12 mutant forming a temperature-sensitive D-serine deaminase. J Bacteriol. 1975 Mar;121(3):1074–1077. doi: 10.1128/jb.121.3.1074-1077.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E., Heincz M. C. Identification and control of synthesis of the dsdC activator protein. J Bacteriol. 1983 Feb;153(2):872–877. doi: 10.1128/jb.153.2.872-877.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E., Heincz M. C. Thermosensitive regulation of D-serine deaminase synthesis in a mutant of Escherichia coli K 12. Mol Gen Genet. 1970;106(4):371–377. doi: 10.1007/BF00324054. [DOI] [PubMed] [Google Scholar]
- McFall E. Mapping of the d-serine deaminase region in Escherichia coli K-12. Genetics. 1967 Jan;55(1):91–99. doi: 10.1093/genetics/55.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E. Role of adenosine 3',5'-cyclic monophosphate and its specific binding protein in the regulation of D-serine deaminase synthesis. J Bacteriol. 1973 Feb;113(2):781–785. doi: 10.1128/jb.113.2.781-785.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E., Runkel L. DNA sequences of the D-serine deaminase control region and N-terminal portion of the structural gene. J Bacteriol. 1983 Jun;154(3):1508–1512. doi: 10.1128/jb.154.3.1508-1512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahel G., Rothstein D. M., Magasanik B. Complex glnA-glnL-glnG operon of Escherichia coli. J Bacteriol. 1982 Apr;150(1):202–213. doi: 10.1128/jb.150.1.202-213.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchaudhuri S., McFall E., Carothers A. M. Specialized transduction of D-serine deaminase genes: formation of lysogens that yield high lambda-d dsd/lambda ratios and formation of a dimeric lambda-d dsd. J Bacteriol. 1976 Aug;127(2):998–1014. doi: 10.1128/jb.127.2.998-1014.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart V., Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J Bacteriol. 1985 Nov;164(2):731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]