Abstract
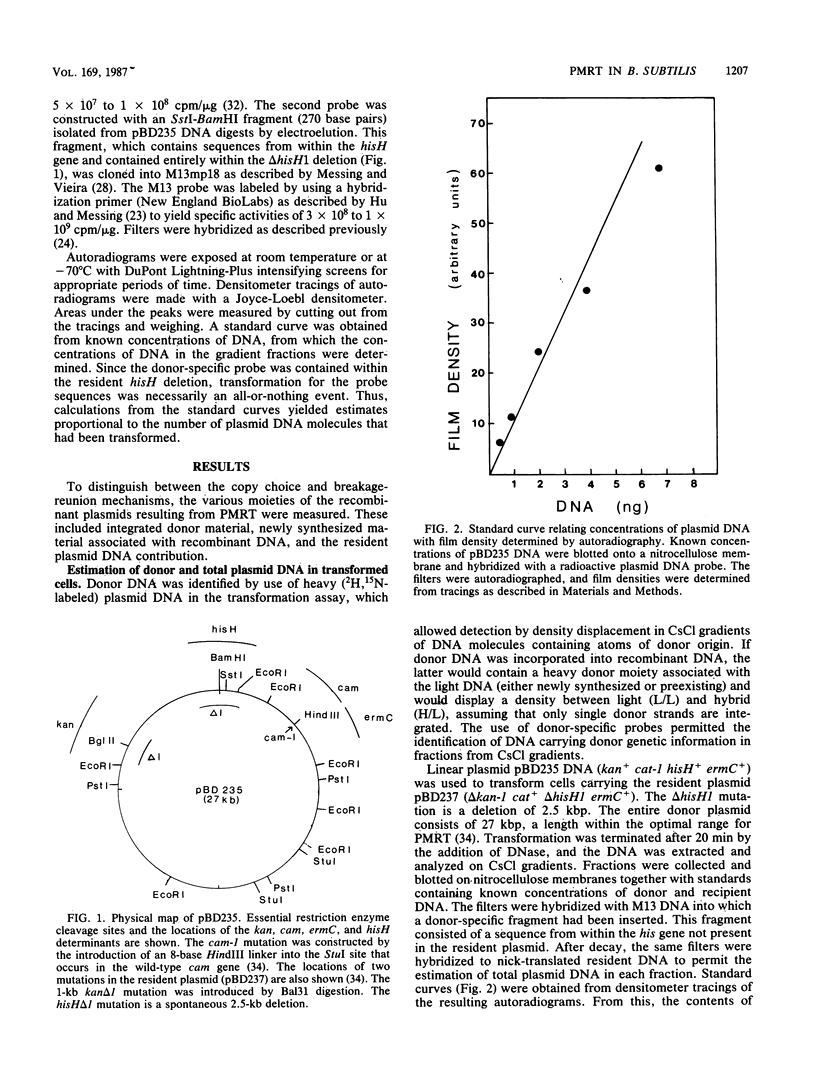
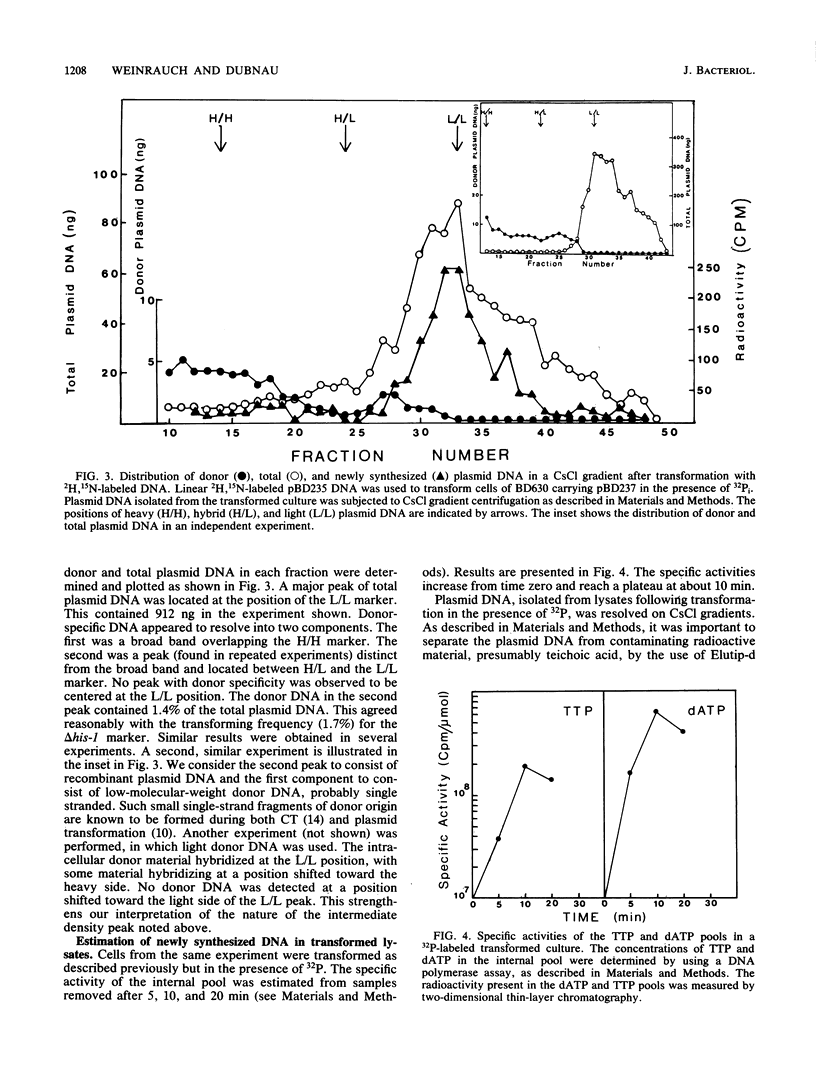
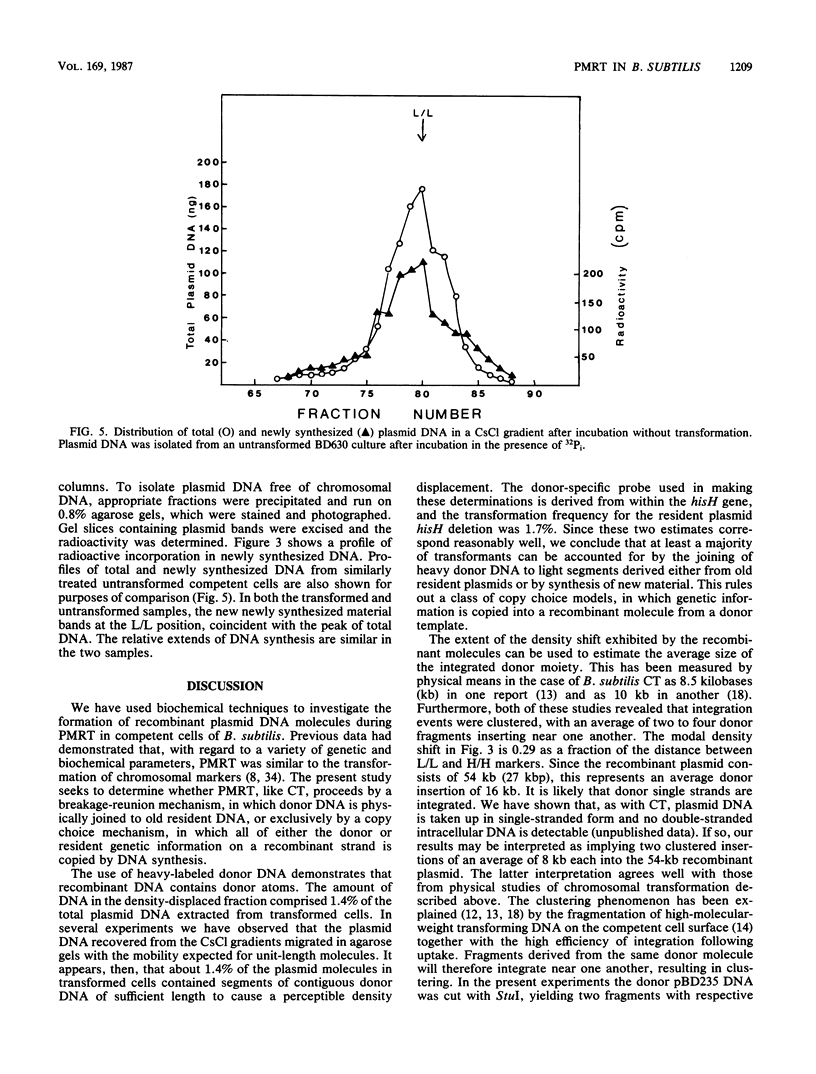
Bacillus subtilis carrying a plasmid which replicates with a copy number of about 1 was transformed with linearized homologous plasmid DNA labeled with the heavy isotopes 2H and 15N, in the presence of 32Pi and 6-(p-hydroxyphenylazo)-uracil to inhibit DNA replication. Plasmid DNA was isolated from the transformed culture and fractionated in cesium chloride density gradients. The distribution of total and donor plasmid DNA was examined, using specific hybridization probes. The synthesis of new DNA, associated with the integration of donor moiety, was also monitored. Donor-specific sequences were present at a density intermediate between that of light and hybrid DNA. This recombinant DNA represented 1.4% of total plasmid DNA. The latter value corresponded well with the transforming activity (1.7%) obtained for the donor marker. Newly synthesized material associated with plasmid DNA at the recombinant density amounted to a minor portion of the recombinant plasmid DNA. These data suggest that, like chromosomal transformation, plasmid marker rescue transformation does not require replication for the integration of donor markers and, also like chromosomal transformation, proceeds by a breakage-reunion mechanism. The extent of donor DNA replacement of recipient DNA per plasmid molecule of 54 kilobases (27 kilobase pairs) was estimated as 16 kilobases.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arwert F., Venema G. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol Gen Genet. 1973;123(2):185–198. doi: 10.1007/BF00267334. [DOI] [PubMed] [Google Scholar]
- Ayad S. R., Barker G. R. The integration of donor and recipient deoxyribonucleic acid during transformation of Bacillus subtilis. Biochem J. 1969 Jun;113(1):167–174. doi: 10.1042/bj1130167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BODMER W. F., GANESAN A. T. BIOCHEMICAL AND GENETIC STUDIES OF INTEGRATION AND RECOMBINATION IN BACILLUS SUBTILIS TRANSFORMATION. Genetics. 1964 Oct;50:717–738. doi: 10.1093/genetics/50.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canosi U., Morelli G., Trautner T. A. The relationship between molecular structure and transformation efficiency of some S. aureus plasmids isolated from B. subtilis. Mol Gen Genet. 1978 Nov 9;166(3):259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- Cashel M., Lazzarini R. A., Kalbacher B. An improved method for thin-layer chromatography of nucleotide mixtures containing 32P-labelled orthophosphate. J Chromatogr. 1969 Mar 11;40(1):103–109. doi: 10.1016/s0021-9673(01)96624-5. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Marker rescue transformation by linear plasmid DNA in Bacillus subtilis. Plasmid. 1979 Oct;2(4):555–571. doi: 10.1016/0147-619x(79)90054-4. [DOI] [PubMed] [Google Scholar]
- Davidoff-Abelson R., Dubnau D. Kinetic analysis of the products of donor deoxyribonucleate in transformed cells of Bacillus subtilis. J Bacteriol. 1973 Oct;116(1):154–162. doi: 10.1128/jb.116.1.154-162.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of products isolated from transformed cells which are derived entirely from donor DNA. J Mol Biol. 1972 Feb 28;64(1):9–29. doi: 10.1016/0022-2836(72)90318-x. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. IV. The endwise attachment and uptake of transforming DNA. J Mol Biol. 1972 Feb 28;64(1):31–46. doi: 10.1016/0022-2836(72)90319-1. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. VI. Non-covalent association of donor and recipient DNA. Mol Gen Genet. 1973 Jan 24;120(2):101–106. doi: 10.1007/BF00267237. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: nonrequirement of deoxyribonucleic acid replication for uptake and integration of transforming deoxyribonucleic acid. J Bacteriol. 1973 Mar;113(3):1512–1514. doi: 10.1128/jb.113.3.1512-1514.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: size and distribution of the integrated donor segments. J Bacteriol. 1972 Aug;111(2):488–494. doi: 10.1128/jb.111.2.488-494.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- FOX M. S., ALLEN M. K. ON THE MECHANISM OF DEOXYRIBONUCLEATE INTEGRATION IN PNEUMOCOCCAL TRANSFORMATION. Proc Natl Acad Sci U S A. 1964 Aug;52:412–419. doi: 10.1073/pnas.52.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornili S. L., Fox M. S. Electron microscope visualization of the products of Bacillus subtilis transformation. J Mol Biol. 1977 Jun 15;113(1):181–191. doi: 10.1016/0022-2836(77)90048-1. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild W. R., Cato A., Jr, Lacks S. Transformation and DNA size: two controlling parameters and the efficiency of the single strand intermediate. Cold Spring Harb Symp Quant Biol. 1968;33:643–645. doi: 10.1101/sqb.1968.033.01.072. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Israeli-Reches M., Weinrauch Y., Dubnau D. Evolutionary relationships of the Bacillus licheniformis macrolide-lincosamide-streptogramin B resistance elements. Mol Gen Genet. 1984;194(3):362–367. doi: 10.1007/BF00425545. [DOI] [PubMed] [Google Scholar]
- Joenje H., Konings W. N., Venema G. Interactions between exogenous deoxyribonucleic acid and membrane vesicles isolated from competent and noncompetent Bacillus subtilis. J Bacteriol. 1975 Mar;121(3):771–776. doi: 10.1128/jb.121.3.771-776.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg U., Skoog L. A method for the determination of dATP and dTTP in picomole amounts. Anal Biochem. 1970 Mar;34:152–160. doi: 10.1016/0003-2697(70)90096-5. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Notani N., Goodgal S. H. On the nature of recombinants formed during transformation in Hemophilus influenzae. J Gen Physiol. 1966 Jul;49(6):197–209. doi: 10.1085/jgp.49.6.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENE J. J., ROMIG W. R. ON THE MECHANISM OF GENETIC RECOMBINATION IN TRANSFORMING BACILLUS SUBTILIS. J Mol Biol. 1964 Jul;9:236–245. doi: 10.1016/s0022-2836(64)80103-0. [DOI] [PubMed] [Google Scholar]
- Randerath K., Randerath E. Ion-exchange thin-layer chromatography. XV. Preparation, properties and applications of paper-like PEI-cellulose sheets. J Chromatogr. 1966 Apr;22(1):110–117. doi: 10.1016/s0021-9673(01)97076-1. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Solter A. W., Handschumacher R. E. A rapid quantitative determination of deoxyribonucleoside triphosphates based on the enzymatic synthesis of DNA. Biochim Biophys Acta. 1969 Feb 18;174(2):585–590. doi: 10.1016/0005-2787(69)90288-3. [DOI] [PubMed] [Google Scholar]
- Weinrauch Y., Dubnau D. Plasmid marker rescue transformation in Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1077–1087. doi: 10.1128/jb.154.3.1077-1087.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos W. M., Venema G., Canosi U., Trautner T. A. Plasmid transformation in Bacillus subtilis: fate of plasmid DNA. Mol Gen Genet. 1981;181(4):424–433. doi: 10.1007/BF00428731. [DOI] [PubMed] [Google Scholar]


