Abstract
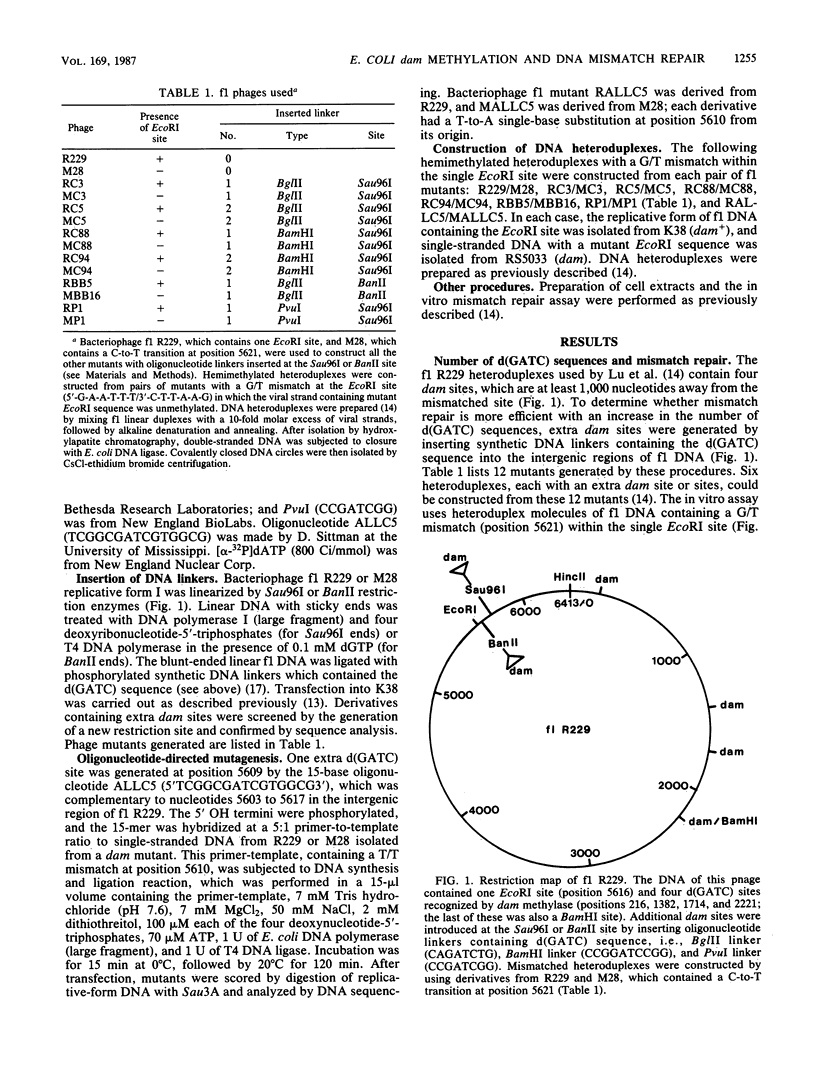
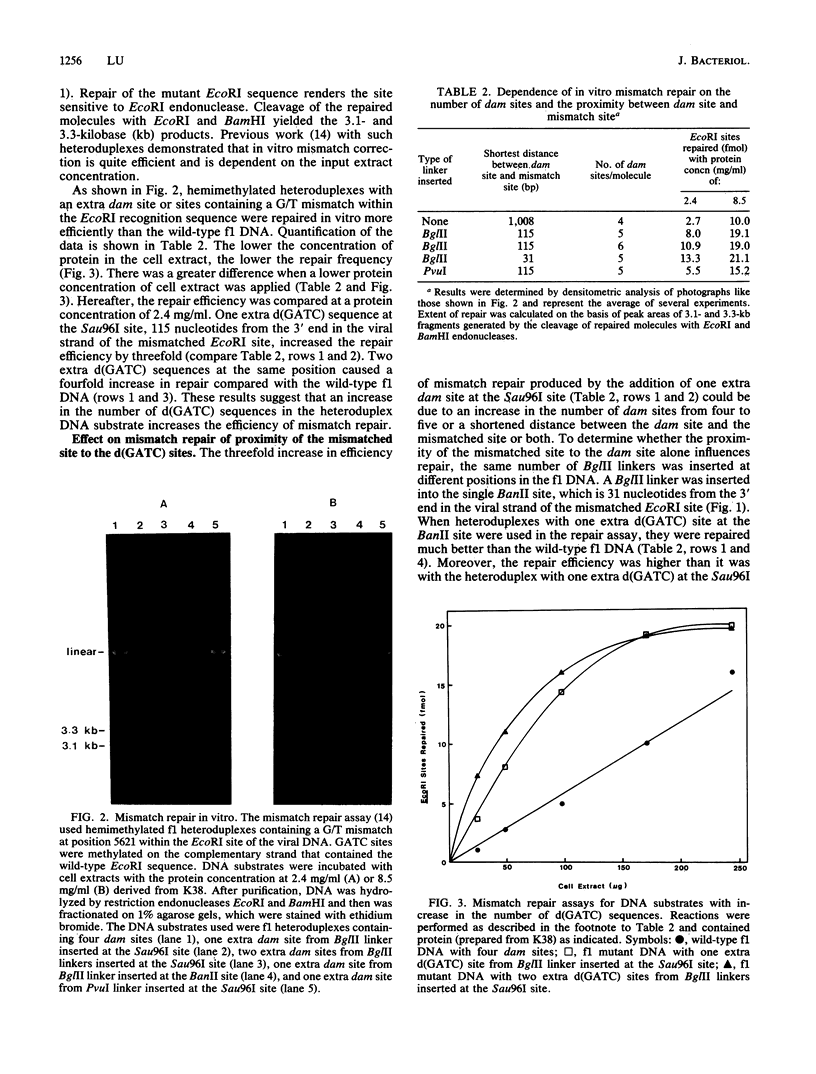
The effect of the number and position of DNA adenine methylation (dam) sites, i.e., d(GATC) sequences, on mismatch repair in Escherichia coli was investigated. The efficiency of repair was measured in an in vitro assay which used an f1 heteroduplex containing a G/T mismatch within the single EcoRI site. Both an increase in the number of dam sites and a shortened distance between dam site and mismatched site increased the efficiency of mismatch repair. The sequences adjacent to d(GATC) also affected the efficiency of methylation-directed mismatch repair. Furthermore, heteroduplexes with one extra dam site located close to either the 5' or 3' end of the excised base increased the repair efficiency to about the same extent. The findings suggest that the mismatch repair pathway has no preferred polarity.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bale A., d'Alarcao M., Marinus M. G. Characterization of DNA adenine methylation mutants of Escherichia coli K12. Mutat Res. 1979 Feb;59(2):157–165. doi: 10.1016/0027-5107(79)90153-2. [DOI] [PubMed] [Google Scholar]
- Bauer J., Krämmer G., Knippers R. Asymmetric repair of bacteriophage T7 heteroduplex DNA. Mol Gen Genet. 1981;181(4):541–547. doi: 10.1007/BF00428750. [DOI] [PubMed] [Google Scholar]
- Boeke J. D. One and two codon insertion mutants of bacteriophage f1. Mol Gen Genet. 1981;181(3):288–291. doi: 10.1007/BF00425599. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E. C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Eichelmann M. C., Lark K. G. Endo R DpnI restriction of Escherichia coli DNA synthesized in vitro. Evidence that the ends of Okazaki pieces are determined by template deoxynucleotide sequence. J Mol Biol. 1977 Dec 15;117(3):621–635. doi: 10.1016/0022-2836(77)90061-4. [DOI] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Hibner U., Alberts B. M. Fidelity of DNA replication catalysed in vitro on a natural DNA template by the T4 bacteriophage multi-enzyme complex. Nature. 1980 May 29;285(5763):300–305. doi: 10.1038/285300a0. [DOI] [PubMed] [Google Scholar]
- Jones M., Wagner R. N-Methyl-N'-nitro-N-nitrosoguanidine sensitivity of E. coli mutants deficient in DNA methylation and mismatch repair. Mol Gen Genet. 1981;184(3):562–563. doi: 10.1007/BF00352542. [DOI] [PubMed] [Google Scholar]
- Konrad E. B. Method for the isolation of Escherichia coli mutants with enhanced recombination between chromosomal duplications. J Bacteriol. 1977 Apr;130(1):167–172. doi: 10.1128/jb.130.1.167-172.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laengle-Rouault F., Maenhaut-Michel G., Radman M. GATC sequence and mismatch repair in Escherichia coli. EMBO J. 1986 Aug;5(8):2009–2013. doi: 10.1002/j.1460-2075.1986.tb04457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Influence of uvrD3, uvrE502, and recL152 mutations on the phenotypes of Escherichia coli K-12 dam mutants. J Bacteriol. 1980 Jan;141(1):223–226. doi: 10.1128/jb.141.1.223-226.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974 May 15;85(2):309–322. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- McGraw B. R., Marinus M. G. Isolation and characterization of Dam+ revertants and suppressor mutations that modify secondary phenotypes of dam-3 strains of Escherichia coli K-12. Mol Gen Genet. 1980;178(2):309–315. doi: 10.1007/BF00270477. [DOI] [PubMed] [Google Scholar]
- Meijer M., Beck E., Hansen F. G., Bergmans H. E., Messer W., von Meyenburg K., Schaller H. Nucleotide sequence of the origin of replication of the Escherichia coli K-12 chromosome. Proc Natl Acad Sci U S A. 1979 Feb;76(2):580–584. doi: 10.1073/pnas.76.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevers P., Spatz H. C. Escherichia coli mutants uvr D and uvr E deficient in gene conversion of lambda-heteroduplexes. Mol Gen Genet. 1975 Aug 27;139(3):233–243. doi: 10.1007/BF00268974. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat Res. 1978 Oct;52(1):11–24. doi: 10.1016/0027-5107(78)90091-x. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Oka A., Sugisaki H., Takanami M., Nishimura A., Yasuda Y., Hirota Y. Nucleotide sequence of Escherichia coli K-12 replication origin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):575–579. doi: 10.1073/pnas.76.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Gruenbaum Y., Urieli-Shoval S., Razin A. Studies on the biological role of DNA methylation: V. The pattern of E.coli DNA methylation. Nucleic Acids Res. 1982 Nov 25;10(22):7247–7259. doi: 10.1093/nar/10.22.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Dohet C., Jones M., Doutriaux M. P., Hutchinson F., Radman M. Involvement of Escherichia coli mismatch repair in DNA replication and recombination. Cold Spring Harb Symp Quant Biol. 1984;49:611–615. doi: 10.1101/sqb.1984.049.01.069. [DOI] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zyskind J. W., Smith D. W. Nucleotide sequence of the Salmonella typhimurium origin of DNA replication. Proc Natl Acad Sci U S A. 1980 May;77(5):2460–2464. doi: 10.1073/pnas.77.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]



