Abstract
The interaction between tomato and its fungal pathogen Cladosporium fulvum complies with the gene-for-gene system, in which specific recognition of fungal proteins by plant genotypes with matching resistance genes results in host resistance. Two proteins, ECP1 and ECP2, secreted by C. fulvum during infection, are required for full virulence of the fungus on tomato. We chose the most important virulence factor, ECP2, for a targeted search for hypersensitive response (HR)-based resistance among a collection of tomato genotypes. By screening with recombinant potato virus X that expresses the Ecp2 gene, we identified four lines that respond with HR toward ECP2. The capacity to recognize ECP2 and induce HR is sufficient to confer resistance in tomato against C. fulvum producing ECP2. Resistance is based on a single dominant gene, which we have designated Cf-ECP2, for resistance to C. fulvum through recognition of ECP2. Accordingly, an Ecp2-minus strain created by gene replacement is pathogenic on Cf-ECP2 plants. However, due to lack of ECP2 the mutant strain is only weakly virulent. All strains of a worldwide collection of C. fulvum strains that were tested were found to produce a HR-inducing ECP2 protein. Because the Cf-ECP2 gene operates through recognition of an important virulence factor, we expect it will confer durable resistance against C. fulvum. A similar targeted approach should allow the discovery of new valuable resistance genes in other pathosystems.
Plant surfaces and intercellular spaces are subjected continuously to potential pathogens. However, individual host plants that exhibit genetic resistance to a particular pathogen often occur in nature. Plant breeders have exploited natural resistance genes extensively via introgression from wild species into high-yielding agronomic cultivars. Nevertheless, this strategy has become less successful as resistance sources are limited and most plant pathogens show remarkable genetic variation leading to the appearance of strains that overcome introgressed genetic resistances (1). Most of the recognition mechanisms in plants remain unrevealed, and the practice of resistance breeding is rather empirical (2). In animal systems, viral vectors are used routinely to deliver antigens to raise antibodies against important pathogens (3–5). Often these antigens are derived from virulence factors of those pathogens. Following a similar approach, plant viruses can be used to deliver virulence factors of plant pathogens to find plants responding with a hypersensitive response (HR). Such plants would likely exhibit a durable type of HR-resistance toward the pathogen from which the virulence factor is originating.
The C. fulvum–tomato (Lycopersicon esculentum) interaction is a well established model system that complies with the gene-for-gene relationship (6). It is also one of the few systems in which there is ample experimental evidence for the involvement of proteinaceous elicitors (avirulence factors) from the pathogen in the induction of active HR-related resistance (7). Many HR-resistant traits have been described in accessions of wild Lycopersicon species and several dominant resistance genes have been introgressed into modern tomato cultivars. HR-mediated resistance in tomato against C. fulvum manifests itself as death of the first cells that come into contact with the penetrating fungus. Further growth of the pathogen is prevented, and the interaction is incompatible. In contrast to resistant plants, susceptible plants do not exhibit HR during interaction with the fungus. Colonization by fungal hyphae occurs through the whole leaf tissue and the interaction is compatible (8). As the development of C. fulvum is restricted to the apoplast of the leaf mesophyll, the elicitors that are secreted by the fungus can be isolated from apoplastic washing fluids (AFs) (9). In this way, two proteinaceous avirulence factors, AVR9 and AVR4, responsible for fungal recognition by host genotypes carrying the matching resistance genes Cf-9 and Cf-4, respectively, have been characterized (10, 11). Two additional fungal extracellular proteins, ECP1 and ECP2, occur abundantly in AF of plants infected by C. fulvum (12, 13). Analysis of mutants of the fungus, in which either the Ecp1 gene or the Ecp2 gene had been deleted, showed that both ECP proteins are virulence factors for C. fulvum. ECP2 is the most important of the two because Ecp2-lacking strains are only weakly pathogenic, exemplified by poor leaf colonization and conidiation (14). By using the potato virus X (PVX) expression system (15), we have identified tomato lines that display HR upon exposure to ECP2. The gene, designated Cf-ECP2, which is responsible for ECP2 recognition confers resistance against C. fulvum on tomato. Because of the crucial role of ECP2 in virulence of the fungus, Cf-ECP2 may prove to be of increased durability and would therefore be valuable in breeding programs aimed at sustainable agriculture.
MATERIALS AND METHODS
Construction of Recombinant PVX∷Ecp2 and PVX Inoculation Procedure.
The chimeric construct for Ecp2 expression and extracellular targeting of the ECP2 protein was obtained by PCR-mediated cloning. The two pairs of oligonucleotides PR1ECP2F (5′-CTTGCCGTGCCCGGAACGCTGGCAACTCGCCC-3′) and ECP2CLA (5′-CGGAAGCTTATCGATCTAGTCATCGTTGGACGGGTTG-3′); and OX10 (5′-CAATCACAGTGTTGGCTTGC-3′) and PR1ECP2R (5′-GTTGCCAGCGTTCCGGGCACGGCAAGAGTGGGATATTAC-3′) were used for PCR with the Ecp2 cDNA and the PVX∷Avr4 construct as templates, respectively (16). After PCR overlap extension and cloning in the PVX vector (15), the final PVX construct carried a chimeric transgene consisting of the sequence coding for the PR-1a plant signal peptide fused to the Ecp2 cDNA encoding the mature protein. This fusion was placed under the control of the PVX coat protein promoter, enabling expression as a subgenomic messenger during virus spread throughout the whole plant. Full-length infectious transcripts were generated in vitro by using the T7 mMESSAGE mMACHINE kit (Ambion, Austin, TX) according to the recommendations of the supplier. The transcripts were inoculated by rubbing onto the leaves of Nicotiana clevelandii plants in presence of carborundum. Ten days after inoculation, infected leaves of N. clevelandii were collected and ground in 50 mM potassium phosphate buffer, pH 7.7, to prepare sap, which contains recombinant virus particles. The sap was used subsequently for inoculation of tomato lines in a similar way as described for N. clevelandii.
C. fulvum Inoculation Procedure.
Suspensions of conidia of C. fulvum were prepared as described (17). In brief, fungal strains were grown on potato dextrose agar plates and conidia were suspended in water by rubbing the sporulating colony. The suspension that was obtained was sprayed onto the lower side of the tomato leaves. Disease symptoms were scored 2–3 weeks after inoculation.
Isolation, Purification, and Immunodetection of ECP2.
Isolation of AFs from susceptible tomato plants inoculated with various C. fulvum strains was performed as described (9). Two weeks after inoculation, infected leaves were collected and water infiltrated in a vacuum chamber, and AF-containing soluble apoplastic compounds was obtained after centrifugation at 3,000 × g for 10 min. Purified ECP2 was obtained from AF of a compatible C. fulvum–tomato interaction by gel filtration on a Sephadex G-50 column (Pharmacia) followed by chromatography on a Resource Q column (Pharmacia) as described by Wubben et al. (13). ECP2-containing fractions were identified with immunodetection by using polyclonal antibodies raised against ECP2 (13). For immunodetection of ECP2, AF containing the extracellular proteins produced in planta by the various strains that were tested, was separated on 15%-polyacrylamide gels containing SDS. The separated proteins were subsequently electro-transferred to Immobilon-P membrane (Millipore), and the blots were incubated with ECP2 polyclonal antibodies.
Crossings.
To obtain three F1 generations, lines 1 and 4 and their common ancestor were crossed to the nonresponsive Moneymaker cultivar, using the latter cultivar as the female parent. These F1 generations were selfed to obtain the F2 generations used in this study.
RESULTS
Identification of Tomato Genotypes that Display HR upon Exposure to ECP2.
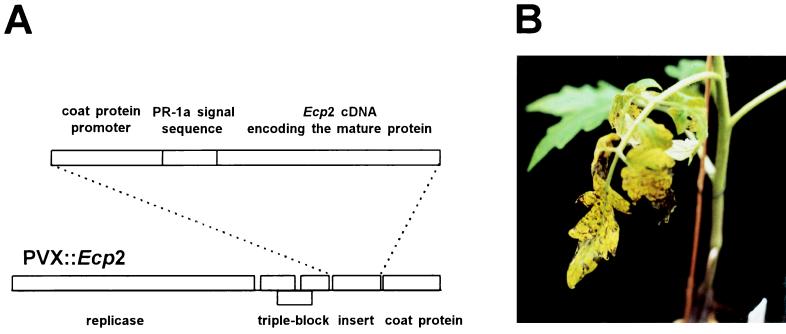
We have screened 21 lines that originated from early breeding programs for resistance against C. fulvum. They had been selected for resistance against the tester strain, race 2.3.4, which overcame all resistance genes, Cf-2, Cf-3, and Cf-4, that had been introgressed at that time (18, 19). The lines that should carry resistance genes different from these three genes were tested for the ability to respond with HR after exposure to ECP2. Screening was carried out by using PVX (15) for systemic production of the ECP2 protein, targeted to the apoplast of virus-infected plants (16), and results were verified with purified native ECP2 protein. PVX∷Ecp2 recombinant virus was obtained by cloning the Ecp2 cDNA encoding the mature ECP2 protein, downstream of the sequence encoding the PR-1a plant signal peptide, under the control of the PVX coat protein promoter (Fig. 1A). Native ECP2 protein was obtained by sequential chromatographic separation of soluble proteins present in AF from a compatible C. fulvum–tomato interaction (Fig. 2A). Four of 21 tomato lines that were tested responded with HR after inoculation with PVX∷Ecp2, as well as after injection with native ECP2. PVX∷Ecp2 inoculation triggered the development of systemic chlorotic and necrotic lesions in leaves of the four lines (Fig. 1B). Upon injection of ECP2, the four lines developed chlorosis followed by necrosis in the center of the injected area within 3 days (Fig. 2B). Inoculation of these four lines with wild-type PVX caused normal mosaic symptoms. The other 17 lines and the control cultivar Moneymaker (lacking any known C. fulvum resistance gene) exhibited normal mosaic symptoms upon inoculation with either PVX∷Ecp2 or wild-type PVX. Systemic HR never was observed in these plants. These results indicate that ECP2 is both necessary and sufficient to induce HR on the four responding lines.
Figure 1.
Construction of the PVX∷Ecp2 derivative and identification of lines responding with HR upon exposure to the ECP2 protein. (A) Schematic map of the PVX∷Ecp2 derivative. (B) Specific HR upon ECP2 presentation. Four-week-old plants of the collection of tomato lines and the control cultivar Moneymaker (lacking any known C. fulvum resistance gene) were inoculated with sap containing PVX∷Ecp2 (16) and symptoms were recorded 10–14 days after inoculation. Control plants of each line were inoculated with sap containing wild-type PVX. All plants gave normal systemic mosaic symptoms upon inoculation with wild-type PVX. Four lines gave systemic chlorosis and necrosis upon inoculation with PVX∷Ecp2. The symptoms on one of these plants, line 1, are shown. Normal mosaic symptoms developed after inoculation with PVX∷Ecp2 on all other plants.
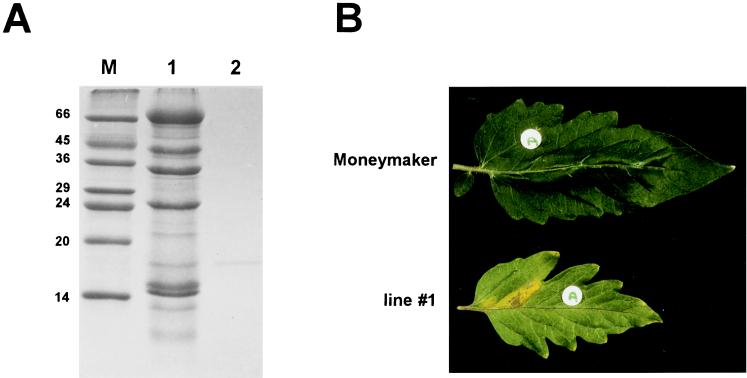
Figure 2.
HR induced after injection of purified ECP2 in a leaflet of line 1. (A) Purification of the ECP2 protein. Lane M contains molecular weight markers, lane 1 contains 50 μl of AF, and lane 2 contains the purified ECP2 protein. (B) HR induced after injection of ECP2 in the apoplast of a leaflet of a line 1 plant (Lower) and absence of HR after injection of ECP2 in the apoplast of a leaflet of the Moneymaker cultivar (Upper). Only the four lines identified by inoculation with PVX∷Ecp2 responded by chlorosis, followed by necrosis in the injected area three days after injection of ECP2; all other lines showed no detectable response after ECP2 injection.
ECP2-Mediated HR Is Determined by a Single Dominant Gene.
The four independent lines that recognize ECP2 have been reported to originate from the same L. pimpinellifolium ancestor and are likely to be four independent introgressions containing the same resistance factor (18, 19). We produced three F1 progenies by crossing line 1, line 4, and their ancestor to the cultivar Moneymaker. F2 progenies were generated to study the heritability of HR upon exposure to ECP2. We inoculated PVX∷Ecp2 onto F1 plants and three F2 populations containing 93, 105, and 85 individuals, respectively, and determined the segregation ratio for HR. All F1 individuals showed HR, and the three F2 populations exhibited a 3:1 segregation for presence to absence of HR after inoculation with PVX∷Ecp2 (Table 1). This demonstrates that one dominant gene determines the capacity to develop HR upon exposure to ECP2 in the ancestor and lines 1 and 4.
Table 1.
Inheritance of HR induced by ECP2
| F1
|
F2
|
||||
|---|---|---|---|---|---|
| HR | Mosaic | HR | Mosaic | χ2 Value | |
| Line 1 × Moneymaker | 4 | 0 | 77 | 16 | 1.13 (P > 0.25) |
| Line 4 × Moneymaker | 5 | 0 | 82 | 23 | 0.27 (P > 0.5) |
| Ancestor × Moneymaker | 4 | 0 | 67 | 18 | 0.25 (P > 0.5) |
Segregation of HR induction by ECP2 in F1 and F2 generations originating from a cross between the HR-displaying line 1, line 4, and their common ancestor with the non-HR-displaying cultivar Moneymaker. Plants were inoculated with PVX∷Ecp2 and scored for systemic HR 2 weeks after inoculation.
ECP2 Recognition Confers Resistance Against C. fulvum.
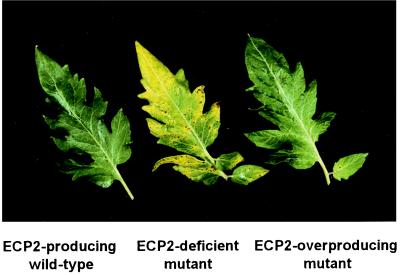
We used isogenic strains of C. fulvum, either with or without the Ecp2 gene, to prove that resistance of these responding lines is solely caused by the fact that the fungus produces ECP2 during infection. We used line 1 as a representative of the four lines. The set of fungal strains tested comprised a wild-type, ECP2-producing strain (race 5), the corresponding isogenic Ecp2-lacking mutant in which the Ecp2 gene had been replaced by an antibiotic resistance cassette, and an ECP2-overproducing strain obtained after retransformation of the Ecp2-lacking mutant with multiple copies of the Ecp2 gene (20). Whereas susceptible Moneymaker plants showed disease symptoms when inoculated with either of the three strains, only the Ecp2-lacking mutant caused disease on line 1 (Fig. 3). Plants of line 1 showed full resistance without any visible disease symptoms after inoculation with the wild-type ECP2-producing strain or the ECP2-overproducing near-isogenic strain. Microscopic examination of these resistant plants confirmed that fungal growth of both the wild-type and the ECP2-overproducing strains is arrested early after penetration of the tomato leaf. The Ecp2-lacking strain colonized to the same low extent the mesophyll of either plants of line 1 or Moneymaker plants (data not shown). The latter observation confirms the role of ECP2 as a virulence factor of C. fulvum on tomato as described before (14). Thus, resistance toward C. fulvum of lines 1 to 4 is solely dependent on recognition of the ECP2 protein, as was expected from the results obtained with the PVX∷Ecp2 experiments. We designate this resistance gene Cf-ECP2 because this single dominant gene confers resistance toward C. fulvum through recognition of the ECP2 protein.
Figure 3.
Resistance of Cf-ECP2 lines is solely dependent on the production of the ECP2 protein by C. fulvum during infection. Plants of line 1 were inoculated with the wild-type ECP2-producing strain (race 5) (Left), the isogenic Ecp2-lacking strain (Center), and the ECP2-overproducing strain (Right). Note that only the plants inoculated with the Ecp2-lacking strain showed symptoms of chlorosis on the upper side of the leaf 20 days after inoculation.
Strains of C. fulvum that Have Been Collected Worldwide Produce a HR-Inducing ECP2 Protein During Infection.
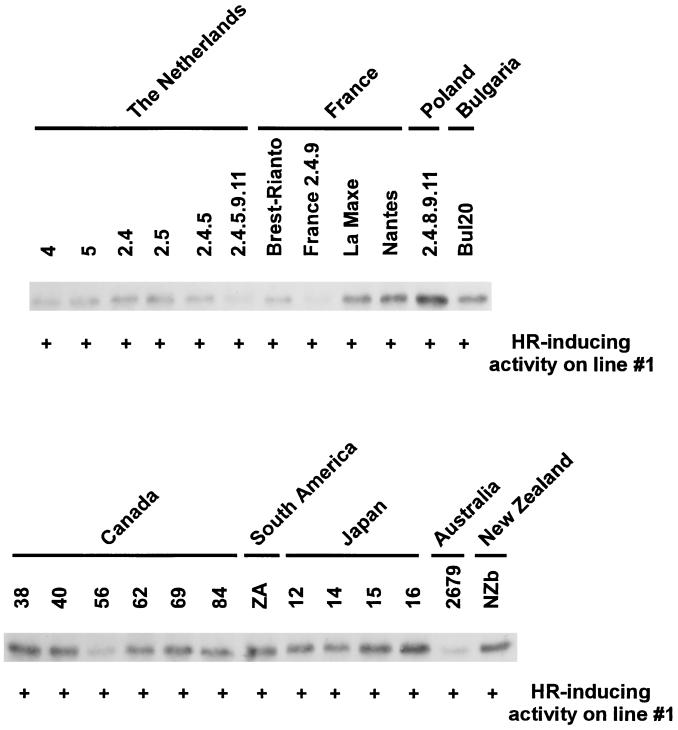
To test the effectiveness of the Cf-ECP2 gene, we analyzed AFs that were obtained from susceptible plants infected by a worldwide collection of 25 strains of C. fulvum, for the presence of the ECP2 protein and its HR-inducing activity. Western blot analysis with polyclonal antibodies raised against ECP2 confirmed the production of ECP2 by all strains (Fig. 4). Differences in accumulation levels of ECP2 are in agreement with accumulation levels of other in planta-secreted proteins of the various C. fulvum strains and reflect their relative aggressiveness. In addition, leaf injection of line 1 with AFs from all 25 strains resulted in the induction of HR, indicating that all strains produce an elicitor-active ECP2 protein (Fig. 4).
Figure 4.
Immunodetection of ECP2 in AFs isolated from cultivar Moneymaker inoculated with 25 strains of C. fulvum obtained from a worldwide collection. A volume of AF containing 4 μg of protein was separated on denaturing polyacrylamide gel, blotted, and incubated with polyclonal antibodies raised against ECP2. ECP2 is detected in each lane. HR-inducing activity of each AF sample was assayed by injecting 5–10 μl into a leaflet of line 1. HR activity visible 3 days after injection is indicated by “+.”
DISCUSSION
Resistance breeding programs are traditionally based on the identification of resistance to a pathogen in wild relatives, followed by several generations of backcrossing to introduce the resistance into elite breeding lines. Initially, inoculations with local and worldwide strains of the pathogen give insight into the effectiveness of the resistance (2). HR-based resistance can be induced by a variety of elicitors originating from the pathogen, of which the importance in pathogenicity or virulence is not known beforehand. If the elicitors are not important pathogenicity or virulence factors, it is likely that the pathogen will easily overcome the resistance either by mutating or losing the encoding gene. Here, we exploited biochemical and molecular data concerning a factor that is important for full virulence of the pathogen and followed a targeted search for plants showing a HR response to this factor.
Frequently, new races of C. fulvum appear that overcome known resistance genes such as Cf-2, Cf-4, Cf-5, and Cf-9. Molecular analysis of strains of C. fulvum that circumvent the Cf-9 resistance gene revealed that complete deletion of the Avr9 gene had occurred in the fungal genome, with the consequence that recognition is avoided as the AVR9 elicitor is no longer produced (21). C. fulvum strains that circumvent the Cf-4 resistance gene exhibit a single point mutation in the Avr4 gene, which causes instability of the encoded product, because no proteins homologous to AVR4 are detected in AF of plants inoculated with these strains (11, 16). So far, no intrinsic biological function has been assigned to the Avr9 and Avr4 genes of C. fulvum. It seems that these genes can be lost without a detectable fitness penalty, so they appear not to encode important factors of pathogenicity or virulence for C. fulvum.
The Cf-ECP2 resistance gene matches an important virulence factor of C. fulvum (14), which is present in all strains of C. fulvum that have been tested up till now. Overexpression of Ecp2 in plants lacking Cf-ECP2 by using the PVX expression system does not give a phenotype, which might be explained by the fact that ECP2 rather suppresses activation of plant defense responses than being a toxic virulence factor. So far, four bacterial and one fungal avirulence gene (avrA and avrE from Pseudomonas syringae pv. tomato, avrRPM1 from Pseudomonas syringae pv. maculicola, avrBs2 from Xanthomonas campestris pv. vesicatoria, and nip1 from Rhynchosporium secalis; refs. 22–25) have been reported to possess a function in virulence. However, these genes were not discovered and isolated based on a targeted search as described here. The matching resistance genes in the host plant are expected to confer durable protection against these pathogens. We do not know whether there are separate domains present in the mature ECP2 protein of 142 amino acids, which are important for its virulence function and for the HR-inducing activity. If there are no separate domains in the ECP2 protein, or if the two separate domains overlap, the fungus cannot circumvent the Cf-ECP2 resistance by mutations in the Ecp2 gene, without a serious decrease in virulence. Altogether, we expect the Cf-ECP2 resistance gene to be efficient and durable in protecting tomato crops against C. fulvum.
Using a viral expression system, such as the one based on PVX, has great potential. It ensures that the plant response is solely due to the protein of which the encoding cDNA has been inserted into the PVX vector. PVX screening can easily be performed with any protein. As in immunization assays used for mammals, the PVX vector presents the protein throughout the infected plant, increasing the amount of responding plant tissue from one single injection site to the whole plant, in which the virus systematically spreads (26). If the heterologous protein expressed by PVX triggers a quick HR response in the host, spread of the virus might be restricted to the inoculated area. In this case, compared with systemic mosaic symptoms caused by wild-type PVX, absence of mosaic symptoms combined with localized necrosis on the inoculated leaves may indicate recognition of the expressed protein. This method allows screening of large populations of wild plants or recombinants for rare individuals that exhibit specific recognition of the protein to which they have been exposed. We are currently testing large collections of wild Lycopersicon species for HR upon exposure to several additional potential virulence factors of C. fulvum. With the availability of several plant viral expression vectors for dicots and monocots, such as PVX, tobacco mosaic virus, cauliflower mosaic virus, tomato golden mosaic virus, cassava latent virus, brome mosaic virus, and barley stripe mosaic virus (15, 27–33), plants other than Solanaceae can be screened for HR-mediated resistance toward these viral vectors expressing pathogenicity or virulence factors produced by economically important plant pathogens.
Acknowledgments
We thank R. Roth for critical reading of the manuscript. R.L. and P.J.G.M.D.W. were supported by the European Community-Human Capital and Mobility program ERBCHRXCT930244. M.H.A.J.J. was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO)/Priority Program Crop Protection. J.P.W.H. was supported by the Associatie van Biotechnologische Onderzoekscholen in Nederland (ABON).
ABBREVIATIONS
- HR
hypersensitive response
- AF
apoplastic fluid
- PVX
potato virus X
References
- 1.Agrios G N. Plant Pathology. San Diego: Academic; 1997. pp. 115–142. [Google Scholar]
- 2.Hayes H K, Immer F R, Smith D C. Methods of Plant Breeding. New York: McGraw–Hill; 1955. pp. 131–151. [Google Scholar]
- 3.Moss B. Proc Natl Acad Sci USA. 1996;93:11341–11348. doi: 10.1073/pnas.93.21.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paoletti E. Proc Natl Acad Sci USA. 1996;93:11349–11353. doi: 10.1073/pnas.93.21.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama N, Maeda K, Mikami T. J Vet Med Sci. 1997;59:311–322. doi: 10.1292/jvms.59.311. [DOI] [PubMed] [Google Scholar]
- 6.Day P R. In: Genetics of Host-Parasite Interaction. Kelman A, Sequeira L, editors. San Francisco: Freeman; 1974. pp. 92–110. [Google Scholar]
- 7.De Wit P J G M. Adv Bot Res. 1995;21:147–185. [Google Scholar]
- 8.De Wit P J G M. In: Biology and Molecular Biology of Plant-Pathogen Interactions. Bailey J, editor. Berlin: Springer; 1986. pp. 149–169. [Google Scholar]
- 9.De Wit P J G M, Spikman G. Physiol Plant Pathol. 1982;21:1–11. [Google Scholar]
- 10.Scholtens-Toma I M J, De Wit P J G M. Physiol Mol Plant Pathol. 1988;33:59–67. [Google Scholar]
- 11.Joosten M H A J, Cozijnsen T J, De Wit P J G M. Nature (London) 1994;367:384–386. doi: 10.1038/367384a0. [DOI] [PubMed] [Google Scholar]
- 12.Joosten M H A J, De Wit P J G M. Physiol Mol Plant Pathol. 1988;33:241–253. [Google Scholar]
- 13.Wubben J P, Joosten M H A J, De Wit P J G M. Mol Plant–Microbe Interact. 1994;7:516–524. doi: 10.1094/mpmi-7-0516. [DOI] [PubMed] [Google Scholar]
- 14.Laugé R, Joosten M H A J, Van den Ackerveken G F J M, Van den Broek H W J, De Wit P J G M. Mol Plant–Microbe Interact. 1997;10:725–734. [Google Scholar]
- 15.Chapman S, Kavanagh T, Baulcombe D. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 16.Joosten M H A J, Vogelsang R, Cozijnsen T J, Verberne M C, De Wit P J G M. Plant Cell. 1997;9:367–379. doi: 10.1105/tpc.9.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Wit P J G M. Neth J Plant Pathol. 1977;83:109–122. [Google Scholar]
- 18.Kanwar J S, Kerr E A, Harney P M. Rep Tomato Genet Coop. 1980;30:20–21. [Google Scholar]
- 19.Kanwar J S, Kerr E A, Harney P M. Rep Tomato Genet Coop. 1980;30:22–23. [Google Scholar]
- 20.Marmeisse R, Van den Ackerveken G F J M, Goosen T, De Wit P J G M, Van den Broek H W J. Curr Genet. 1994;26:245–250. doi: 10.1007/BF00309555. [DOI] [PubMed] [Google Scholar]
- 21.Van Kan J A L, Van den Ackerveken G F J M, De Wit P J G M. Mol Plant–Microbe Interact. 1991;4:52–59. doi: 10.1094/mpmi-4-052. [DOI] [PubMed] [Google Scholar]
- 22.Lorang J M, Shen H, Kobayashi D, Cooksey D, Keen N T. Mol Plant–Microbe Interact. 1994;7:508–515. [Google Scholar]
- 23.Ritter C, Dangl J L. Mol Plant–Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 24.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohe M, Gierlich A, Hermann H, Hahn M, Schmidt B, Rosahl S, Knogge W. EMBO J. 1995;14:4168–4177. doi: 10.1002/j.1460-2075.1995.tb00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammond-Kosack K E, Staskawicz B J, Jones J D G, Baulcombe D C. Mol Plant–Microbe Interact. 1995;8:181–185. [Google Scholar]
- 27.Takamatsu N, Ishikawa M, Meshi T, Okada Y. EMBO J. 1987;6:307–311. doi: 10.1002/j.1460-2075.1987.tb04755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donson J, Kearney C M, Hilf M E, Dawnson W O. Proc Natl Acad Sci USA. 1991;88:7204–7208. doi: 10.1073/pnas.88.16.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brisson N, Paszkowski J, Penswick J R, Gronenborn B, Potrykus I, Hohn T. Nature (London) 1984;310:511–514. [Google Scholar]
- 30.Hayes R J, Petty I T D, Coutts R H A, Buck K W. Nature (London) 1988;334:179–182. [Google Scholar]
- 31.Ward A, Etessami P, Stanley J. EMBO J. 1988;7:1583–1587. doi: 10.1002/j.1460-2075.1988.tb02983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.French R, Janda M, Ahlquist P. Science. 1986;231:1294–1297. doi: 10.1126/science.231.4743.1294. [DOI] [PubMed] [Google Scholar]
- 33.Joshi R L, Joshi V, Ow D W. EMBO J. 1990;9:2663–2669. doi: 10.1002/j.1460-2075.1990.tb07451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]