Abstract
The proton–sucrose symporter that mediates phloem loading is a key component of assimilate partitioning in many higher plants. Previous biochemical investigations showed that a diethyl pyrocarbonate-sensitive histidine residue is at or near the substrate-binding site of the symporter. Among the proton–sucrose symporters cloned to date, only the histidine residue at position 65 of AtSUC1 from Arabidopsis thaliana is conserved across species. To test whether His-65 is involved in the transport reaction, we have used site-directed mutagenesis and functional expression in yeast to determine the significance of this residue in the reaction mechanism. Symporters with mutations at His-65 exhibited a range of activities; for example, the H65C mutant resulted in the complete loss of transport capacity, whereas H65Q was almost as active as wild type. Surprisingly, the H65K and H65R symporters transport sucrose at significantly higher rates (increased Vmax) than the wild-type symporter, suggesting His-65 may be associated with a rate-limiting step in the transport reaction. RNA gel blot and protein blot analyses showed that, with the exception of H65C, the variation in transport activity was not because of alterations in steady-state levels of mRNA or symporter protein. Significantly, those symporters with substitutions of His-65 that remained transport competent were no longer sensitive to inactivation by diethyl pyrocarbonate, demonstrating that this is the inhibitor-sensitive histidine residue. Taken together with our previous results, these data show that His-65 is involved in sucrose binding, and increased rates of transport implicate this region of the protein in the transport reaction.
Assimilate partitioning is a fundamental process that allows plants to function as multicellular organisms. In general, oxidized forms of carbon and nitrogen are reductively assimilated in photosynthetic tissues, and then sugars and amino acids are transported in the phloem cells of the plant’s vascular system to the heterotrophic organs. In many plants, sucrose is the principal form of translocated photoassimilate and, therefore, it is a central metabolite in plant growth and development. Moreover, sucrose accumulation in the phloem is a pivotal step in partitioning because it produces a large osmotic potential that generates the positive hydrostatic pressure that drives long-distance transport. A key contributor in this system of resource allocation is the proton–sucrose symporter that transports sucrose into the phloem against a large concentration difference (1, 2).
The transport properties and bioenergetics of the proton–sucrose symporter were initially described in intact tissue systems, and then more detailed analysis was achieved by using purified plasma membrane vesicles and imposed proton electrochemical potential differences (see ref. 2 for review). The symporter is a secondary active carrier that couples sucrose translocation across the plasma membrane to the protonmotive force generated by the H+-pumping ATPase. The transporter is specific for sucrose with an apparent Km of 1 mM (3–5). The transport reaction is electrogenic and the proton:sucrose stoichiometry is 1:1 (6–8).
The symporter is inhibited by a variety of compounds, including p-chloromercuribenzenesulfonic acid (3, 9, 10), diethyl pyrocarbonate (DEPC) (3, 11), cytochalasin B (12), and forskolin (11, 12). The chemical properties of such inhibitors, as well as their molecular interactions with the carrier, often provide information about amino acid residues that are involved in substrate binding and translocation. For example, DEPC-dependent inactivation of the sucrose symporter is the result of chemical modification of a histidine residue, and kinetic analysis of the inactivation reaction showed that the sensitive histidine is at, or conformationally linked to, the sucrose-binding site (12). Moreover, it was shown that the DEPC-sensitive residue is accessible from the outside face of the plasma membrane, suggesting it is located in a region of the symporter that is exposed to the extracellular solution (12). These results implicate a histidine residue in substrate binding.
The sucrose symporter was recently cloned from spinach and potato by using functional complementation of a yeast invertase mutant (13, 14). Additional examples have also been cloned from Arabidopsis thaliana (AtSUC1) (15), Plantago major (16), and sugar beet (unpublished data). These cDNAs encode proteins with calculated molecular masses of ≈50–55 kDa, they are very similar to one another, with 55–84% amino acid identity, and hydropathy analysis suggests that they contain 12 transmembrane domains. Significantly, the transport properties of the heterologously expressed carrier in yeast cells, or membrane vesicles isolated from them, are similar to those described in plant membrane vesicles (2, 13, 14, 16). Additional experiments showed that these transporters are involved in phloem loading, because they are expressed in a phloem-specific manner (14, 17–19) and antisense expression of the potato clone resulted in a stunted plant phenotype that is consistent with a block in assimilate partitioning (20). In spite of these exciting advances in describing the biochemical and molecular properties of the symporter, however, little is known about its structure and function relationships.
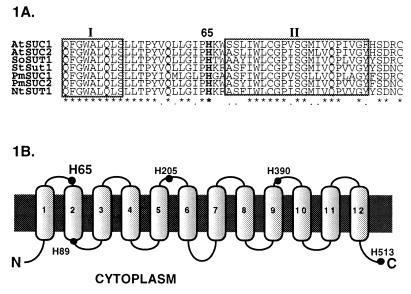
As a first step toward defining the amino acid residues and protein domains that play key roles in the transport reaction, we have used site-directed mutagenesis to determine the role of the DEPC-sensitive histidine residue in sucrose transport. Among the five histidine residues in the deduced amino acid sequence of AtSUC1, only His-65 is conserved in all proton–sucrose symporters cloned to date (Fig. 1A). In the results reported here, we show that this is the DEPC-sensitive residue in the sucrose symporter and that some mutations in this residue result in dramatic changes in transport activity.
Figure 1.
(A) Partial alignment of the proton–sucrose symporters. The boxed regions represent a portion of the first and the whole second transmembrane domains of AtSUC1. The His-65 residue is the only histidine of the symporter conserved across species. (B) Topological model of AtSUC1. The putative two-dimensional model of the AtSUC1 consists of 12 transmembrane α-helices. The filled circles indicate the putative locations of the five histidine residues of AtSUC1 in the context of the predicted topology.
MATERIALS AND METHODS
Expression of AtSUC1 in Yeast.
An EcoRI fragment containing the full-length cDNA of AtSUC1 was cloned into pNEV-E, a yeast/Escherichia coli shuttle vector containing the Saccharomyces cerevisiae PMA1 promoter (15). The resulting plasmid containing the cDNA insert in the sense orientation was used to transform the yeast mutant DBY2615 (his4-539 lys2-801 ura3-52 SUC2Δ), which is deficient in utilizing extracellular sucrose (21). Uracil-independent transformants were identified on SD minimal medium (2% glucose/0.67% yeast nitrogen base without amino acids/0.08% complete synthetic amino acid mixture minus uracil).
Single-stranded DNA Isolation and Site-Directed Mutagenesis.
AtSUC1 cDNA was cloned into pBluescript II (−) KS (Stratagene) and the resulting plasmid, pBS-AtSUC1, was used to transform CJ236 [dut-1 ung-1 thi1 relA-1 pCJ105 (chloramphenicol-resistant)], an E. coli strain incapable of removing uracil misincorporated into DNA during replication. Single-stranded DNA extraction and site-directed mutagenesis of His-65 residue of AtSUC1 were based on the protocol from the Muta-Gene M13 In Vitro Mutagenesis Kit (Bio-Rad). Briefly, a 40-mer oligonucleotide that was degenerate for His-65 (underlined trimer), CGTTCAACTCCTCGGGATCCCTNNNAAATGGTCATCTCTC, was used to prime synthesis of a second strand of the AtSUC1 plasmid. The double-stranded plasmid was then transformed into an E. coli strain that degrades the uracil-containing strand of plasmid DNA that carries the wild-type SUC1 insert. The primer also included an exogenous BamHI restriction site (underlined hexamer) with a silent mutation just three nucleotides 5′ of the His-65 codon. Plasmids extracted from E. coli transformants were screened for incorporation of the 40-mer by BamHI digestion. Mutant codons were determined by DNA sequencing with a specific primer 5′ to the His-65 codon by using the dideoxynucleotide chain-termination method (22) with Sequenase version 2.0 (United States Biochemical).
Transformation of S. cerevisiae.
Competent cells of DBY2615 were prepared by harvesting cells grown in SD to midlogarithmic phase and were suspended in 1 M sorbitol at ≈1 × 1010 cells per ml. Aliquots (50 μl) of the cell suspension were mixed with 0.5 μg of plasmid DNA. Transformation was performed by electroporation, and transformed cells were selected on SD plates containing 1 M sorbitol.
Transport Assays.
Cells were grown to logarithmic phase, washed, and resuspended at 150–200 mg of cells per ml, in a sugar-free NSC medium (0.67% yeast nitrogen base without amino acids, 0.08% complete synthetic amino acid mixture minus uracil) at pH 5.2. Transport assays were initiated by diluting 20 μl of cells into 0.2 ml of NSC medium containing 10 mM glucose, 1 mM unlabeled sucrose, and 0.2 μCi (1 μCi = 37 kBq) of U-14C-labeled sucrose. An aliquot of suspended cells was delivered onto a micropore filter (0.45 μm pore size) at desired time points and the transport solution was aspirated into a flask by vacuum filtration. The cells were washed three times with 2 ml of ice-cold water to remove extracellular radiolabel. Isotope accumulation in the cells was determined by scintillation spectroscopy. All transport experiments were repeated at least three times. For pH dependence of sucrose transport experiments, cells were treated the same way but resuspended in 25 mM sodium phosphate buffer of pH 5, 6, 7, or 8.
RNA Gel Blot Analysis.
A 50-ml culture of yeast grown in SD medium was harvested at saturation and RNA was extracted by using a bead beater in the presence of equal volumes of phenol and extraction buffer (100 mM Tris⋅HCl, pH 8.0/10 mM EDTA/1% SDS). The homogenate was briefly centrifuged to remove cell debris and the supernatant was extracted with an equal volume of chloroform. RNA was pelleted with ¼ vol of LiCl then resuspended and treated with DNase I at 37°C for 30 min. Total RNA was purified with an additional extraction with phenol/chloroform followed by precipitation with ethanol. In RNA gel blot analysis, 30 μg of total RNA from each construct was electrophoresed on a 1.5% agarose gel containing formaldehyde and transferred to a nylon membrane (23). The probe was prepared with random hexamer-primed [α-32P]dATP labeling of the EcoRI fragment of AtSUC1. Equal loading was evaluated by staining the membrane with methylene blue in 5% acetic acid.
Microsome Preparation and Protein Gel Blot Analysis.
Transgenic yeast cells were harvested at midlogarithmic phase and broken with glass beads. Microsomes were isolated according to standard protocols (24). Microsomal proteins of each transgenic yeast line were analyzed on SDS/10% PAGE (25) and transferred to a poly(vinylidene difluoride) (PVDF) membrane (Millipore) with a Semi-Dry Transfer Cell (Bio-Rad). Antibody was detected by using the protocol described in ref. 26. A partially purified anti-AtSUC1 antiserum was kindly provided by R. Stadler (19).
DEPC Inhibition.
Yeast were grown and harvested at midlogarithmic phase. Cells were washed in 4°C H2O and then in 25 mM sodium phosphate buffer (pH 5.5). Equal volumes of cells and DEPC solutions were mixed to initiate inactivation reactions. Reactions were terminated after 30 sec by adding 6-fold larger volumes of 500 μM l-histidine in the sodium phosphate buffer followed by mixing and centrifugation. The spent solution was removed and the cells were washed a second time. The washed cells were resuspended in the sodium phosphate buffer and used in transport experiments.
RESULTS
There are five histidine residues in the AtSUC1 protein: His-65, His-89, His-205, His-390, and His-513. Sequence alignments between the spinach (13), potato (14), Plantago major (16), Arabidopsis thaliana (15), and sugar beet (unpublished data) clones showed that, among these histidines, only His-65 is conserved in each porter (Fig. 1A). Moreover, current models of the topology of the symporter suggest that His-65 is located in the first extracellular loop near the second transmembrane domain of the protein (Fig. 1B). This location is consistent with earlier biochemical analysis showing that the DEPC-sensitive residue is accessible from the extracellular face of the plasma membrane (12). On the basis of these observations, we used site-directed mutagenesis to explore the role of His-65 in sucrose transport.
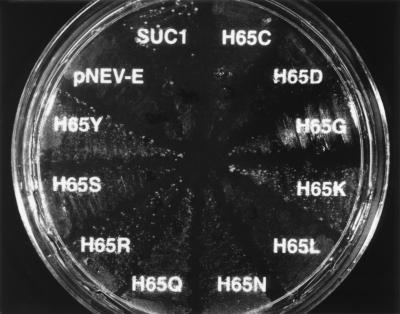
A series of single amino acid substitutions at His-65 were generated that include Cys, Asp, Gly, Lys, Leu, Asn, Gln, Arg, Ser, and Tyr. To investigate the impact of these substitutions on transport activity, each mutant form of suc1 was ligated into a yeast expression vector and used to transform DBY2615, a yeast mutant deficient in cell wall invertase that also lacks sucrose transport activity (21), to score for functional complementation. A sucrose-limiting medium (1 mM sucrose as the sole carbon source) was established where yeast cells expressing wild-type SUC1 grow slowly, but insert-free vector controls do not. Several patterns of growth were observed in DBY2615 cells transformed with the mutant forms of SUC1 (Fig. 2). Cells expressing SUC1 mutants with Arg or Lys substitutions grew faster than cells with the wild-type symporter. Symporters with Asn and Gln substitutions allowed growth at about the same rate as wild type; and Gly-, Ser-, and Tyr-substituted porters supported slower growth. Leu and Asp substitutions gave small slow-growing colonies, and the Cys-substituted symporter did not complement growth (Fig. 2). These results support the hypothesis that His-65 is an important residue in sucrose transport.
Figure 2.
Growth complementation of S. cerevisiae DBY2615 by the wild-type and His-65 → Xaa mutant proteins of AtSUC1 on 1 mM sucrose as the sole carbon source. Sucrose at 1 mM was used as the sole carbon source in minimal medium to differentiate the growth of yeast expressing the wild-type AtSUC1 from insert-free vector controls and transgenic yeast carrying His-65 → Xaa mutant constructs.
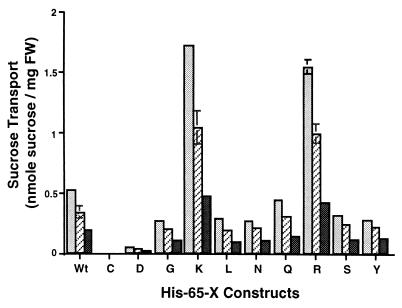
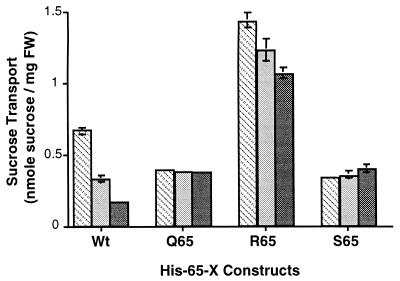
Different rates of growth suggested the modified symporters exhibited divergent rates of sucrose transport. We investigated that possibility by measuring the rate of sucrose transport into the expressing yeast cells (Fig. 3). Those results were complementary to our observations regarding growth rates. For example, Arg- and Lys-substituted symporters transport sucrose at much higher rates than does wild type, as much as 310%. These results suggest His-65 substitutions may alter the basic transport properties of the symporter.
Figure 3.
Histogram of sucrose transport by transgenic S. cerevisiae expressing the wild-type AtSUC1 or Suc1 mutants of residue 65. Sucrose transport (4-min time points) by transgenic yeast was assayed in the presence of 0.2 mM (darkly shaded bars), 0.5 mM (hatched bars), and 1 mM (lightly shaded bars) sucrose. FW, fresh weight.
If His-65 is involved in the translocation reaction of the sucrose symporter, we would expect changes in the kinetics of sucrose transport by the substituted mutants and, indeed, several mutants exhibited changes in Vmax (Gly- and Tyr-65), Km (Leu-, Asn-, and Ser-65), or both (Asp-, Lys-, Gln-, and Arg-65) (Table 1). Although examples of altered Vmax or Km were observed in these modified transporters, it appears that Vmax had the most significant impact on transport activity. For example, the higher Vmax values seen in the Lys- and Arg-substituted porters (14- and 8-fold, respectively) had a more pronounced effect on transport activity than did changes in Km. Likewise, lower Vmax values were observed in the slow porters, even when they also had higher apparent affinities—i.e., lower Km values.
Table 1.
Kinetic analysis of sucrose transport by His-65 mutants of AtSUC1 expressed in S. cerevisiae DBY2615
| H65X | Vmax, nmol/mg⋅min | Km, mM |
|---|---|---|
| Wild type | 0.177 | 0.500 |
| H65D | 0.021 | 0.367 |
| H65G | 0.104 | 0.526 |
| H65K | 2.52 | 4.26 |
| H65L | 0.129 | 0.794 |
| H65N | 0.134 | 0.800 |
| H65Q | 0.285 | 1.36 |
| H65R | 1.52 | 2.67 |
| H65S | 0.148 | 0.754 |
| H65Y | 0.101 | 0.410 |
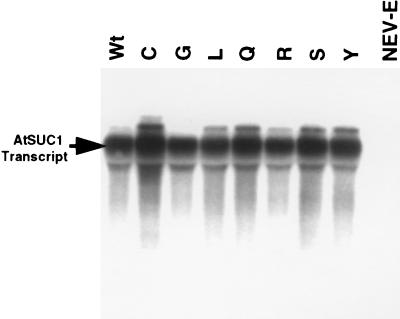
Altered Vmax for transport activity could reflect differential levels of gene expression. Therefore, we measured the steady-state levels of AtSUC1 mRNA and protein in both wild type and mutants. RNA gel blot analysis of total RNA from each construct expressed in DBY2615 showed that all of the transgenic yeast expressed the SUC1 mRNA to the same level (Fig. 4). Note that the steady-state levels of Cys-65 mRNA did not diverge from those of the other mutants, indicating that the absence of function was not because of lack of message.
Figure 4.
RNA gel blot analysis of the transgenic yeast expressing the wild-type AtSUC1 or SUC1 with mutations at residue 65. The blot was probed with 32P-labeled AtSUC1 cDNA. Equal loading of total RNA was examined by staining the blot with methylene blue (data not shown).
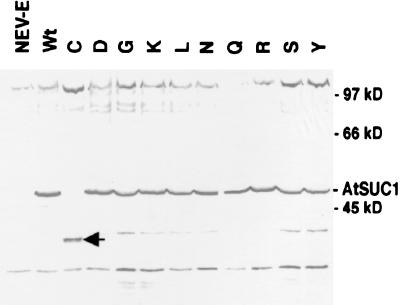
Protein gel blot analysis of the SUC1 mutants showed about the same level of the symporter protein at approximately 47 kDa (Fig. 5), with the exception of the H65C mutant. These results suggest that the variation in transport activity results from changes in protein structure and function versus alterations in protein abundance. To examine whether the Cys-65 mutant protein was not integrated in the membrane fraction, the soluble fraction of Cys-65 complemented cells was also analyzed. No evidence of AtSUC1 Cys-65 protein was found in the soluble fraction of the cells, and there was no intact protein present in the total microsomal fraction after longer detection by the antibody against AtSUC1 (data not shown). The presence of smaller protein bands in the Cys-65 lane that are not visible in the lanes of wild type or the other mutant forms of the symporter suggests that the Cys-65 protein is synthesized and then degraded (Fig. 5).
Figure 5.
Protein gel blot analysis of the transgenic yeast expressing insert-free vector (NEV-E), wild-type AtSUC1, or SUC1 with mutations at residue 65. SUC1 protein was detected at approximately 47 kDa by partially purified anti-SUC1 antibody. The arrowhead indicates lower molecular mass bands of Cys-65 AtSUC1 detected by the antibody.
We examined DEPC-dependent inhibition of sucrose transport by AtSUC1 to test whether His-65 is the DEPC-sensitive residue of the transporter. Transgenic yeast cells expressing the Gln-65, Arg-65, or Ser-65 form of SUC1 were insensitive to DEPC inhibition compared with the wild-type transporter (Fig. 6). This result suggests that His-65 is the inhibitor-sensitive histidine in AtSUC1 and, moreover, that it is the only histidine residue whose chemical modification affects the transport reaction.
Figure 6.
Concentration-dependent inactivation of sucrose transport in DEPC-treated yeast expressing His-65-Xaa constructs of AtSUC1. Sucrose transport was measured after cells were treated for 30 sec with 0 μM (hatched bars), 50 μM (lightly shaded bars), or 500 μM (darkly shaded bars) DEPC. Transport was measure for 4 min in 1 mM sucrose.
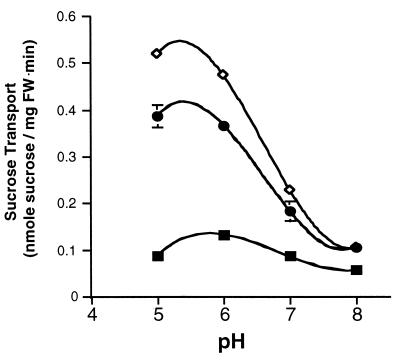
To see whether mutations at His-65 had an effect on apparent proton binding, we examined the pH dependence of sucrose transport in wild-type and mutant forms of the symporter. As shown in Fig. 7, wild-type proteins had highest activity at pH 6.0, and activity declined at elevated pH. Mutants with Lys-65 and Arg-65 exhibited a similar pattern of pH dependence for sucrose transport.
Figure 7.
pH dependence of sucrose transport by the wild type and His-65-Xaa AtSUC1 mutants in DBY2615. Sucrose transport was measured in the presence of 1 mM sucrose with 10 mM glucose in 25 mM sodium phosphate buffer at pH 5.0, 6.0, 7.0, and 8.0. ■, Wild type; ♦, Lys-65; and •, Arg-65 AtSUC1.
DISCUSSION
In the results reported here we show that His-65 plays an important role in the sucrose transport activity of the Arabidopsis thaliana AtSUC1 proton–sucrose symporter. Earlier biochemical analysis of symporter sensitivity to DEPC suggested that a histidine residue is at or near the substrate-binding site (12). Using site-directed mutants of the symporter, we demonstrate that His-65 is the DEPC-sensitive residue (Fig. 6). In addition, we show that the transport activity of mutant symporters varies widely, depending on the chemical properties of the amino acid residues substituted for His-65. For example, the Asp-65 mutant was only 15% as active as the wild-type symporter, whereas Lys-65 and Arg-65 mutants were 3- to 14-fold more active than wild type (Fig. 3, Table 1). Enhanced transport activity was the result of increased Vmax, suggesting that changes in His-65 alter a rate-limiting step in the transport reaction (Table 1). RNA gel blot and Western blot analyses showed that variation in transport activity, with the exception of the cysteine substitution, was not the result of altered transcript or protein expression levels (Figs. 4 and 5). Taken together, these results strongly support a role for His-65 in the transport reaction of the sucrose symporter.
Histidine residues have been implicated in the transport mechanisms of other proton-coupled transporters. His-322 of the E. coli lactose permease was initially suggested to participate in the proton translocation pathway and also substrate recognition (27, 28). Likewise, His-376 of LacS of Streptococcus thermophilus was implicated in sugar recognition for the cotransport of protons and galactosides (29). More recently, His-322 of lactose permease has been proposed to play a role in hydrogen bonding to a critical acidic residue (Glu-269) during the transport reaction without direct participation in proton translocation (30). Although such results focus additional attention on His-65, it is important to note that those histidine residues may not play the same role in their respective proteins as His-65 does in the sucrose symporter. For example, the H322R mutation of LacY lost protonmotive-force-coupling, and the H376Q mutation of LacS resulted in complete loss of transport function, whereas in AtSUC1 H65Q and H65R mutations lead to equal or higher transport rates than that of the wild-type symporter. Moreover, the DEPC-sensitive histidine(s) of lactose permease is not directly involved in substrate binding (31). In contrast, substrate protection experiments have shown that His-65 is at, or at least conformationally linked to, the substrate-binding site of the sucrose symporter (ref. 12; Fig. 6).
Does His-65 participate in proton translocation? Although the pK of the imidazole ring of histidine residues (≈6.0) may be in an appropriate range for reversible proton binding, our results with basic amino acid substitutions suggest that this is not the function of His-65. Both lysine- and arginine-substituted symporters exhibited enhanced transport activity. The pK values of these residues are approximately 10 and 12, respectively. Thus, it is hard to imagine how these residues could participate in a reversible protonation reaction. Although there is precedence for extreme perturbations in the pK values of amino acid side chains, these usually occur when the reactive groups are buried inside very nonpolar regions of the protein (32). In this case, hydropathy analysis and earlier biochemical evidence (12) both suggest that His-65 is near the outside face of the plasma membrane. This is a very polar and acidic environment in plant cells. Moreover, the Lys-65 and Arg-65 mutants display acidic pH optima that are very similar to that of the wild-type symporter (Fig. 7), further complicating their potential participation in a reversible protonation reaction. Therefore, we conclude that His-65 is not directly involved in the proton translocation reaction.
Previous investigations of substrate interaction with the symporter suggested the 3-, 4-, and 6-hydroxyls of the glucosyl moiety of sucrose are important for substrate binding, and the fructosyl half contains a hydrophobic surface that is also critical for substrate recognition (33–35). Given the size differences between the imidazole ring of His-65 and the basic amino acids, it is difficult to picture how their positive charges could hydrogen bond with one of the important hydroxyl groups of sucrose. On the other hand, one might speculate that positive charge plays a role in a salt bridge in this region of the transporter that is important for function. However, that would be hard to reconcile with the wild-type transport activity observed when neutral amino acids were substituted for histidine (Fig. 3). Likewise, enhanced transport activity in the presence of positively charged residues argues against a hydrophobic interaction with sucrose.
Even if His-65 does not interact directly with sucrose, it can still play an important role in defining the substrate-binding site and/or the translocation pathway. For example, His-322 of lactose permease is irreplaceable with respect to proton-coupled transport, although current evidence suggests it does not directly participate in proton translocation or substrate binding. In the most recent model of energy coupling in lactose permease, His-322 plays a crucial role in stabilizing positional relationships by hydrogen bonding to an acidic amino acid residue (Glu-269) in one of the six transmembrane domains that have been implicated in the translocation pathway (30).
We suggest His-65 plays an important role in defining the translocation pathway and participating in the reaction mechanism of the sucrose symporter. This proposal is consistent with substrate protection from inactivation by DEPC (12). This hypothesis is also compatible with the changes observed in Km and Vmax between the various substituted mutants. If His-65 were simply blocking substrate access to the binding pocket, we would expect to see changes only in Km in our mutant symporters. Yet, significant enhancement in Vmax was observed with basic amino acid substitutions, suggesting that a rate-limiting step has been altered in the translocation process. Thus, we conclude His-65 is also a participant in the transport reaction.
The hypothesis that His-65 is involved in the transport reaction is supported by the high degree of sequence conservation in this region of the cloned symporters. Many sugar transporters are members of the major facilitator superfamily (MFS) of carriers that share significant levels of amino acid similarity and predicted protein topology (36–39). Although the sucrose symporters may be related members of this family, they are distinct from both hexose and disaccharide transporters. For example, whereas His-65 is found in all sucrose symporters cloned to date, a comparable histidine is not conserved in proton-coupled hexose or disaccharide transporters in the MFS. However, the HUP1 glucose transporter from Chlorella kessleri does contain a histidine residue in this region (His-73), and arginine-substituted mutants of that residue do not alter transport activity (40). The hydrophilic loop between transmembrane domains I and II exhibits the highest level of sequence conservation among all the loop regions of the sucrose symporters. Moreover, this region appears to be unique to the sucrose symporters, supporting the hypothesis that His-65 is part of the translocation pathway and further suggesting that this domain plays a role in defining substrate specificity. Significantly, the first external loop of the Chlorella glucose symporter was also found to be involved in substrate recognition (41), and the first transmembrane domain of lactose permease was recently implicated in substrate recognition (42).
The proton–sucrose symporter is a key participant in phloem translocation and assimilate partitioning. In the results reported here we provide evidence that His-65 is a critical residue in the transport reaction mediated by the symporter. We are currently characterizing several random mutations in the symporter that were identified in a screen for symporters exhibiting altered transport activity. Not surprisingly, one of those random mutants turned out to be an independent isolation of a transporter mutated at His-65 (data not shown). Taking such results together with the results reported here, we hope to identify enough essential residues to allow us to build an experimentally based model of the structural domains of the symporter that participate in the transport reaction. In addition, we recently described a previously unknown signaling pathway that controls the expression level and transport activity of the sucrose symporter (43). That discovery lends additional significance to defining the functional domains of this vital transporter.
Acknowledgments
We thank Dr. Norbert Sauer (Lehrstuhl Botanik II, Friedrich Alexander Universität Erlangen, Erlangen, Germany) for kindly providing the cDNA clone of AtSUC1 and Dr. Colin Wraight for helpful comments on the manuscript.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: DEPC, diethyl pyrocarbonate.
References
- 1.Giaquinta R T. Annu Rev Plant Physiol. 1983;34:347–387. [Google Scholar]
- 2.Bush D R. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:513–542. [Google Scholar]
- 3.Bush D R. Plant Physiol. 1989;89:1318–1323. doi: 10.1104/pp.89.4.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemoine R E, Delrot S. FEBS Lett. 1989;249:129–133. [Google Scholar]
- 5.Buckhout T J. Planta. 1989;178:393–399. doi: 10.1007/BF00391867. [DOI] [PubMed] [Google Scholar]
- 6.Bush D R. Plant Physiol. 1990;93:1590–1596. doi: 10.1104/pp.93.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slone J H, Buckhout T J. Planta. 1991;183:584–589. doi: 10.1007/BF00194280. [DOI] [PubMed] [Google Scholar]
- 8.Boorer K J, Loo D D F, Frommer W B, Wright E M. J Biol Chem. 1996;271:25139–25144. doi: 10.1074/jbc.271.41.25139. [DOI] [PubMed] [Google Scholar]
- 9.Giaquinta R T. Plant Physiol. 1976;57:872–875. doi: 10.1104/pp.57.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams L E, Nelson S J, Hall J L. Planta. 1990;182:540–545. doi: 10.1007/BF02341029. [DOI] [PubMed] [Google Scholar]
- 11.Williams L E, Nelson S J, Hall J L. Planta. 1992;186:541–550. doi: 10.1007/BF00198034. [DOI] [PubMed] [Google Scholar]
- 12.Bush D R. Arch Biochem Biophys. 1993;307:355–360. doi: 10.1006/abbi.1993.1600. [DOI] [PubMed] [Google Scholar]
- 13.Riesmeier J W, Willmitzer L, Frommer W B. EMBO J. 1992;11:4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riesmeier J W, Hirner B, Frommer W B. Plant Cell. 1993;5:1591–1598. doi: 10.1105/tpc.5.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer N, Stolz J. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- 16.Gahrtz M, Stolz J, Sauer N. Plant J. 1994;6:697–706. doi: 10.1046/j.1365-313x.1994.6050697.x. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn C, Franceschi V R, Schulz A, Lemoine R, Frommer W B. Science. 1997;275:1298–1300. doi: 10.1126/science.275.5304.1298. [DOI] [PubMed] [Google Scholar]
- 18.Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N. Plant Cell. 1995;7:1545–1554. doi: 10.1105/tpc.7.10.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stadler R, Sauer N. Bot Acta. 1996;109:299–306. [Google Scholar]
- 20.Kuhn C, Quick W P, Schulz A, Riesmeier J W, Sonnewald U, Frommer W B. Plant Cell Environ. 1996;19:1115–1123. [Google Scholar]
- 21.Kaiser C A, Botstein D. Mol Cell Biol. 1986;6:2382–2391. doi: 10.1128/mcb.6.7.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Serrano R. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Chiou T J, Bush D R. Plant Physiol. 1996;110:511–520. doi: 10.1104/pp.110.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaback H R. Biochemistry. 1987;26:2071–2076. doi: 10.1021/bi00382a001. [DOI] [PubMed] [Google Scholar]
- 28.Poolman B, Royer T J, Mainzer S E, Schmidt B F. J Bacteriol. 1989;171:244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poolman B, Modderman R, Reizer J. J Biol Chem. 1992;267:9150–9157. [PubMed] [Google Scholar]
- 30.Kaback H R. Proc Natl Acad Sci USA. 1997;94:5539–5543. doi: 10.1073/pnas.94.11.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Padan E, Patel L, Kaback H R. Proc Natl Acad Sci USA. 1979;76:6221–6225. doi: 10.1073/pnas.76.12.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fersht A. Enzyme Structure and Mechanism. New York: Freeman; 1985. [Google Scholar]
- 33.Hitz W D, Card P J, Ripp K G. J Biol Chem. 1986;261:11986–11991. [PubMed] [Google Scholar]
- 34.Delrot S, Roques N, Descotes G, Mentech J. Plant Physiol Biochem. 1991;29:25–29. [Google Scholar]
- 35.Hecht R, Slone J H, Buckhout T J, Hitz W D, Vanderwoude W J. Plant Physiol. 1992;99:439–444. doi: 10.1104/pp.99.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffith J K, Baker M E, Duncan R A, Page M G P, Skurrar R A, Paulsen I T, Chater K F, Baldwin S A, Henderson P J F. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- 37.Henderson P J F. Curr Opin Cell Biol. 1993;5:708–721. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 38.Marger M D, Saier M H. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 39.Goswitz V C, Brooker R J. Protein Sci. 1995;4:534–537. doi: 10.1002/pro.5560040319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caspari T, Stadler R, Sauer N, Tanner W. J Biol Chem. 1994;269:3498–3502. [PubMed] [Google Scholar]
- 41.Will A, Tanner W. FEBS Lett. 1996;381:127–130. doi: 10.1016/0014-5793(96)00097-x. [DOI] [PubMed] [Google Scholar]
- 42.Varela M F, Brooker R J, Wilson T H. J Bacteriol. 1997;179:5570–5573. doi: 10.1128/jb.179.17.5570-5573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiou T-J, Bush D R. Proc Natl Acad Sci USA. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]