Abstract
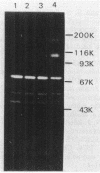
The gene encoding beta-amylase was cloned from Bacillus polymyxa 72 into Escherichia coli HB101 by inserting HindIII-generated DNA fragments into the HindIII site of pBR322. The 4.8-kilobase insert was shown to direct the synthesis of beta-amylase. A 1.8-kilobase AccI-AccI fragment of the donor strain DNA was sufficient for the beta-amylase synthesis. Homologous DNA was found by Southern blot analysis to be present only in B. polymyxa 72 and not in other bacteria such as E. coli or B. subtilis. B. polymyxa, as well as E. coli harboring the cloned DNA, was found to produce enzymatically active fragments of beta-amylases (70,000, 56,000, or 58,000, and 42,000 daltons), which were detected in situ by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Nucleotide sequence analysis of the cloned 3.1-kilobase DNA revealed that it contains one open reading frame of 2,808 nucleotides without a translational stop codon. The deduced amino acid sequence for these 2,808 nucleotides encoding a secretory precursor of the beta-amylase protein is 936 amino acids including a signal peptide of 33 or 35 residues at its amino-terminal end. The existence of a beta-amylase of larger than 100,000 daltons, which was predicted on the basis of the results of nucleotide sequence analysis of the gene, was confirmed by examining culture supernatants after various cultivation periods. It existed only transiently during cultivation, but the multiform beta-amylases described above existed for a long time. The large beta-amylase (approximately 160,000 daltons) existed for longer in the presence of a protease inhibitor such as chymostatin, suggesting that proteolytic cleavage is the cause of the formation of multiform beta-amylases.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Collmer A., Schoedel C., Roeder D. L., Ried J. L., Rissler J. F. Molecular cloning in Escherichia coli of Erwinia chrysanthemi genes encoding multiple forms of pectate lyase. J Bacteriol. 1985 Mar;161(3):913–920. doi: 10.1128/jb.161.3.913-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg F. On the primary structure of amylases. FEBS Lett. 1983 Feb 21;152(2):139–140. doi: 10.1016/0014-5793(83)80365-2. [DOI] [PubMed] [Google Scholar]
- Friedberg F., Rhodes C. Cloning and characterization of the beta-amylase gene from Bacillus polymyxa. J Bacteriol. 1986 Mar;165(3):819–824. doi: 10.1128/jb.165.3.819-824.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien N. H., Fleet G. H. Separation and characterization of six (1 leads to 3)-beta-glucanases from Saccharomyces cerevisiae. J Bacteriol. 1983 Dec;156(3):1204–1213. doi: 10.1128/jb.156.3.1204-1213.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Negoro S., Okada H. Molecular basis of isozyme formation of beta-galactosidases in Bacillus stearothermophilus: isolation of two beta-galactosidase genes, bgaA and bgaB. J Bacteriol. 1984 Oct;160(1):9–14. doi: 10.1128/jb.160.1.9-14.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. General Biochemical Characterization of Thermostable Extracellular beta-Amylase from Clostridium thermosulfurogenes. Appl Environ Microbiol. 1985 May;49(5):1162–1167. doi: 10.1128/aem.49.5.1162-1167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara H., Sasaki T., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a thermophilic alpha-amylase gene: homology between prokaryotic and eukaryotic alpha-amylases at the active sites. J Biochem. 1985 Jul;98(1):95–103. doi: 10.1093/oxfordjournals.jbchem.a135279. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Renaturation of enzymes after polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate. J Biol Chem. 1980 Aug 10;255(15):7467–7473. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrain C., Stalon V., Glansdorff N. Escherichia coli ornithine carbamolytransferase isoenzymes: evolutionary significance and the isolation of lambdaargF and lambdaargI transducing bacteriophages. J Bacteriol. 1976 Oct;128(1):35–38. doi: 10.1128/jb.128.1.35-38.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J. J. Characterization of Bacillus polymyxa amylase as an exo-acting(1 leads to 4)-alpha-D-glucan maltohydrolase. FEBS Lett. 1974 Sep 15;46(1):1–4. doi: 10.1016/0014-5793(74)80321-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mézes P. S., Blacher R. W., Lampen J. O. Processing of Bacillus cereus 569/H beta-lactamase I in Escherichia coli and Bacillus subtilis. J Biol Chem. 1985 Jan 25;260(2):1218–1223. [PubMed] [Google Scholar]
- Ramakrishna N., Dubnau E., Smith I. The complete DNA sequence and regulatory regions of the Bacillus licheniformis spoOH gene. Nucleic Acids Res. 1984 Feb 24;12(4):1779–1790. doi: 10.1093/nar/12.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kuroda M., Sakaguchi K. Isolation and characterization of four plasmids from Bacillus subtilis. J Bacteriol. 1977 Mar;129(3):1487–1494. doi: 10.1128/jb.129.3.1487-1494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A., Tsukagoshi N., Udaka S. Reassembly in vitro of hexagonal surface arrays in a protein-producing bacterium, Bacillus brevis 47. J Bacteriol. 1982 Sep;151(3):1485–1497. doi: 10.1128/jb.151.3.1485-1497.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]