Abstract
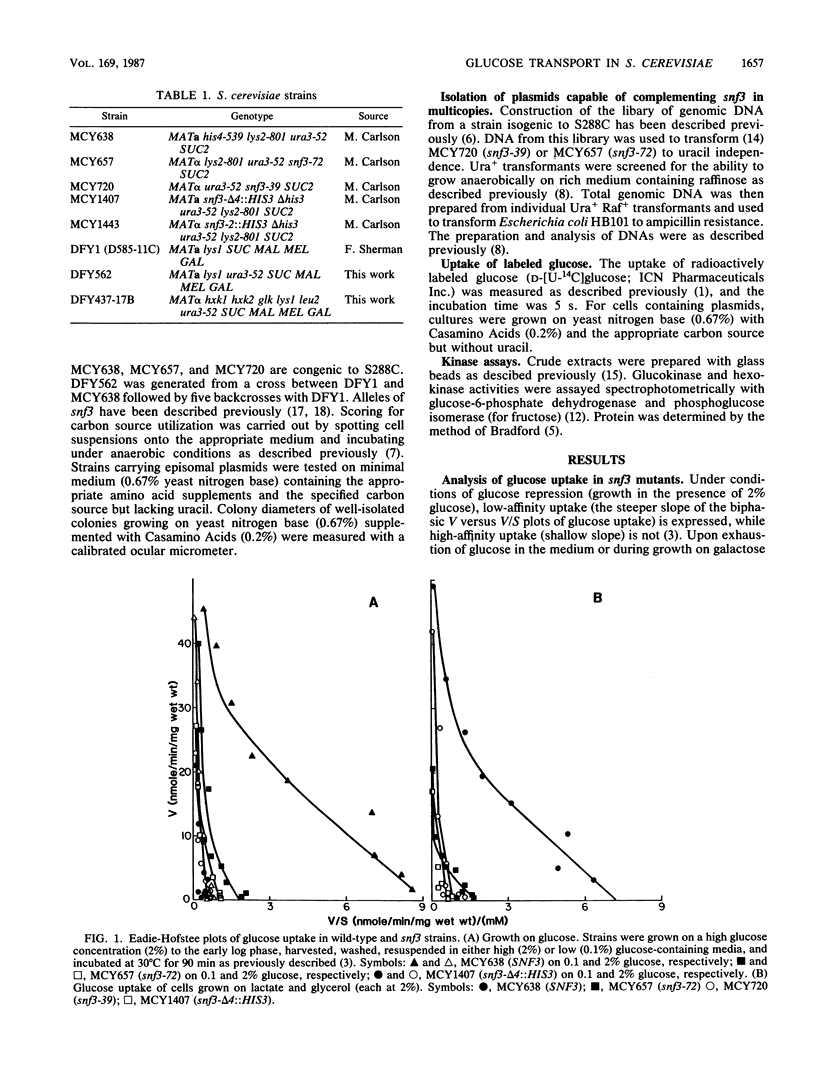
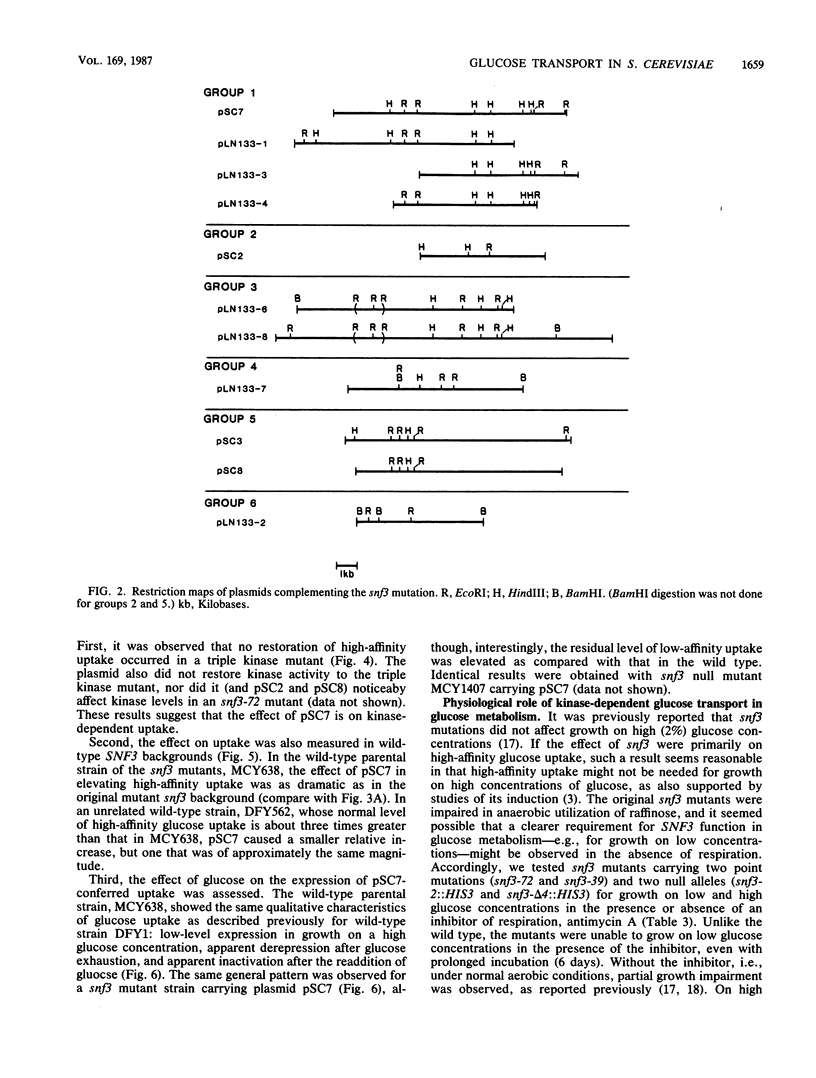
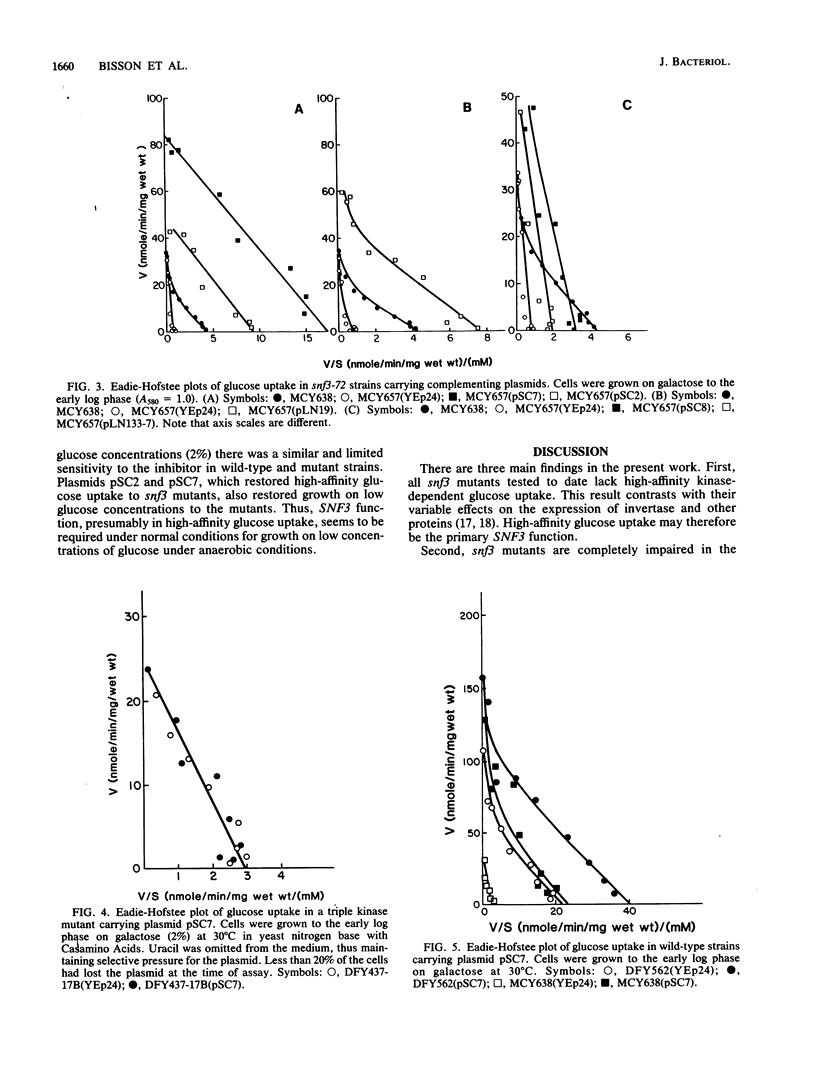
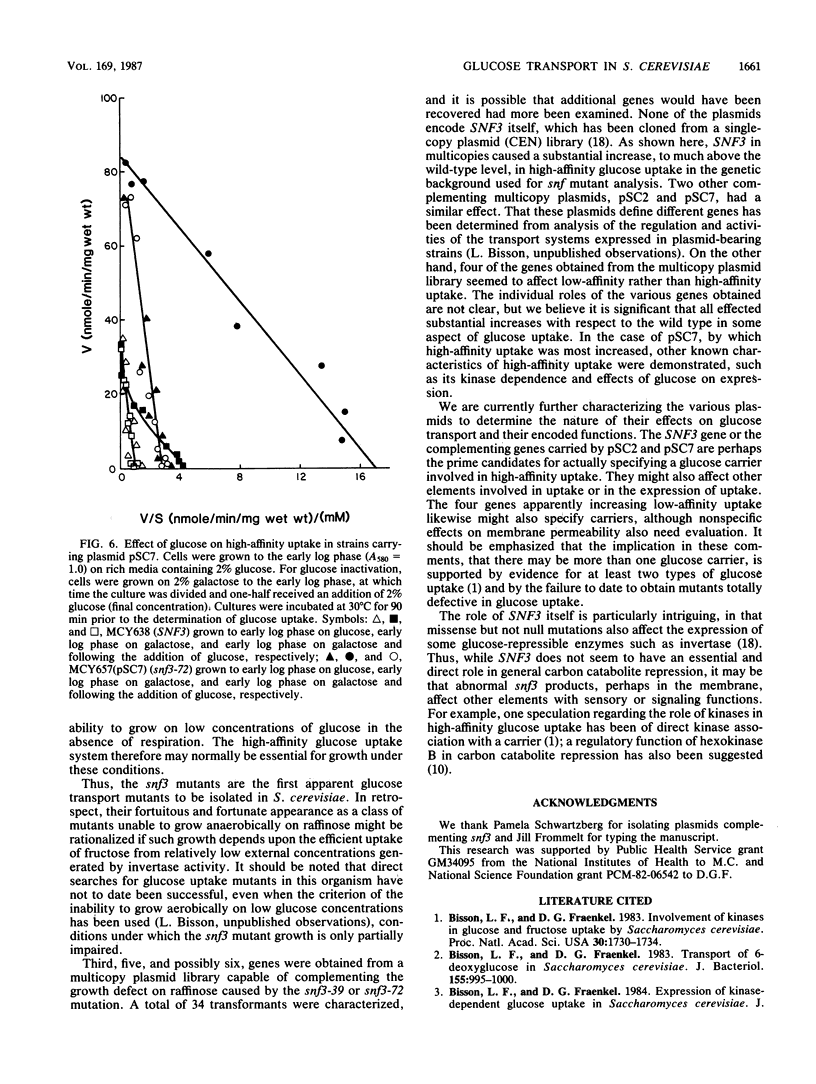
Glucose uptake mutants have not been previously obtained in Saccharomyces cerevisiae, possibly because there seem to be at least two transport systems, of low and high affinities. We showed that snf3 (sucrose nonfermenting) mutants did not express high-affinity glucose uptake. Furthermore, their growth was completely impaired on low concentrations of glucose in the presence of antimycin A (which blocks respiration). Several genes which complemented the original snf3 gene were obtained on multicopy plasmids. Some of them, as well as plasmid-carried SNF3 itself, conferred a substantial increase in high-affinity glucose uptake in both snf3 and wild-type hosts. The effects of glucose on the expression of such a plasmid-determined high-affinity uptake resembled those in the wild type. Other genes complementing snf3 seemed to cause an increase in low-affinity glucose uptake. We suggest that SNF3 may function specifically in high-affinity glucose uptake, which is needed under some conditions of growth on low glucose concentrations. SNF3 itself or the other complementing genes may specify components of the glucose uptake system.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bisson L. F., Fraenkel D. G. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1730–1734. doi: 10.1073/pnas.80.6.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson L. F., Fraenkel D. G. Transport of 6-deoxyglucose in Saccharomyces cerevisiae. J Bacteriol. 1983 Sep;155(3):995–1000. doi: 10.1128/jb.155.3.995-1000.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jan;4(1):49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Fröhlich K. U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984 Apr;158(1):29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo J. M., Clifton D., Fraenkel D. G. Yeast hexokinase mutants. J Biol Chem. 1977 Jul 10;252(13):4443–4444. [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki G., Fraenkel D. G. Cloning of yeast glycolysis genes by complementation. Biochem Biophys Res Commun. 1982 Oct 15;108(3):1107–1122. doi: 10.1016/0006-291x(82)92114-3. [DOI] [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Resistance to 2-deoxyglucose in yeast: a direct selection of mutants lacking glucose-phosphorylating enzymes. Mol Gen Genet. 1977 Dec 9;157(3):297–300. doi: 10.1007/BF00268666. [DOI] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984 Dec;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Schwartzberg P., Reid R., Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986 Nov;6(11):3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]


