Abstract
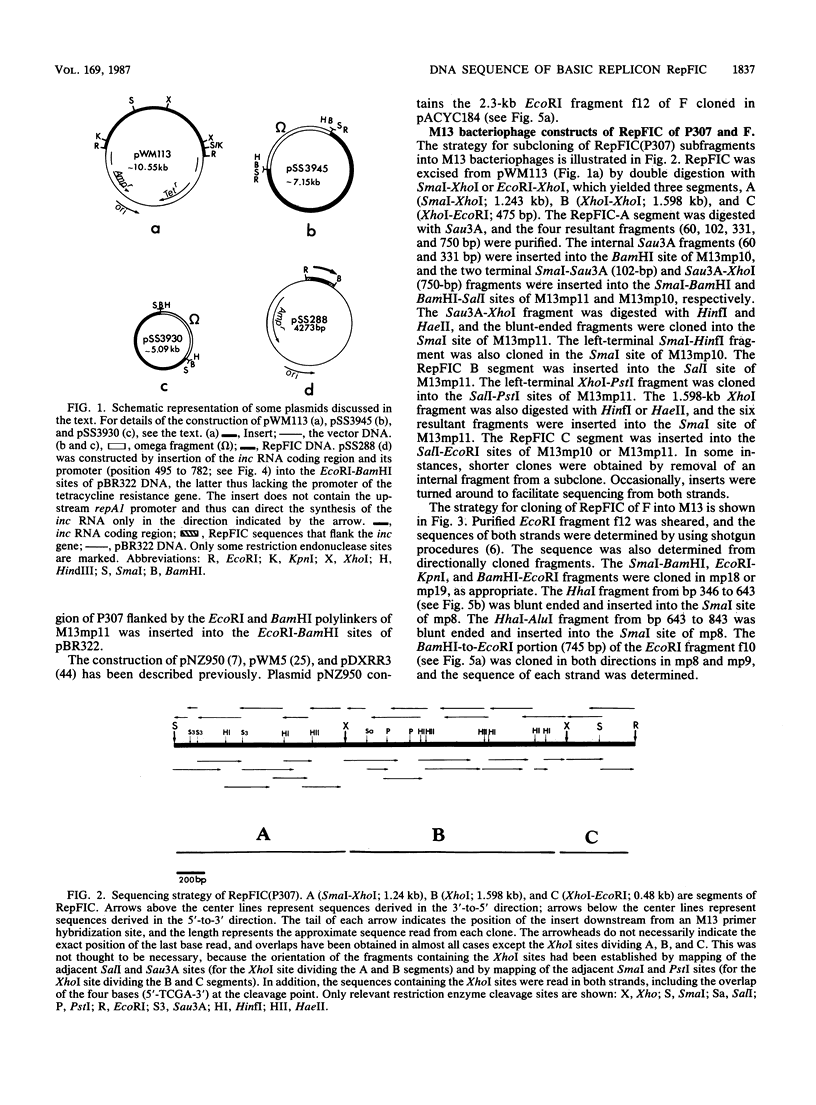
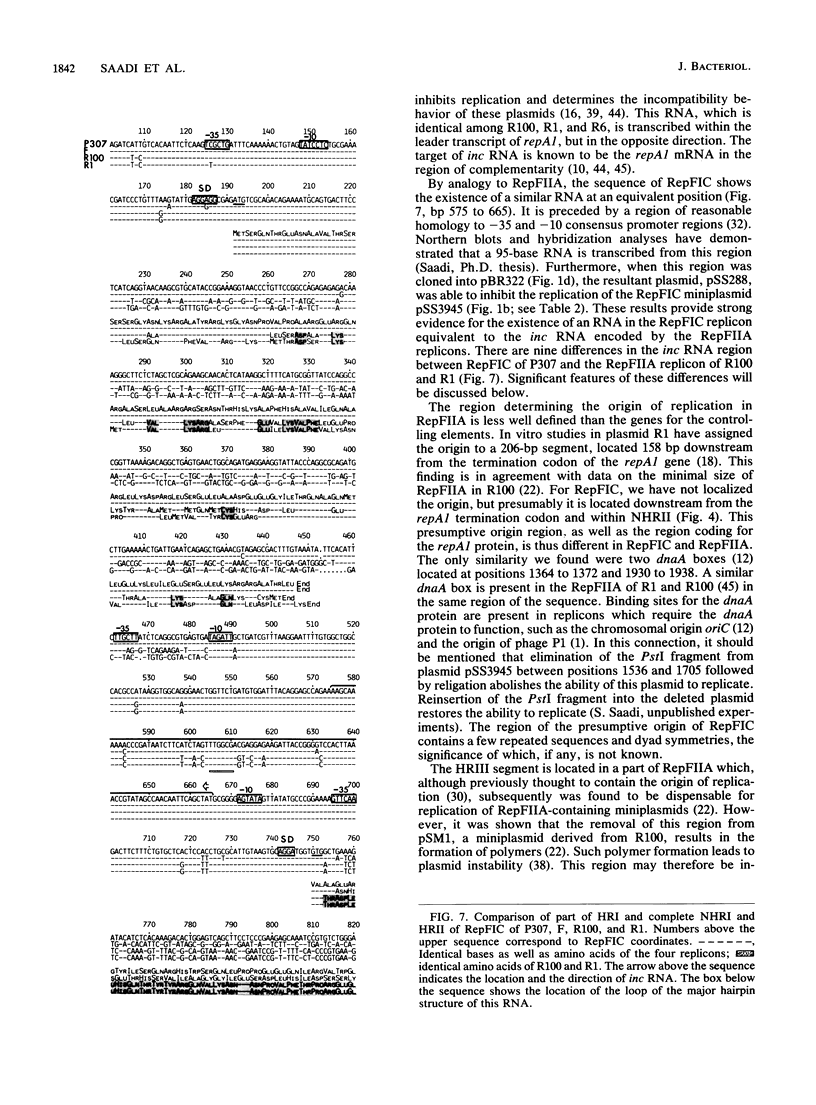
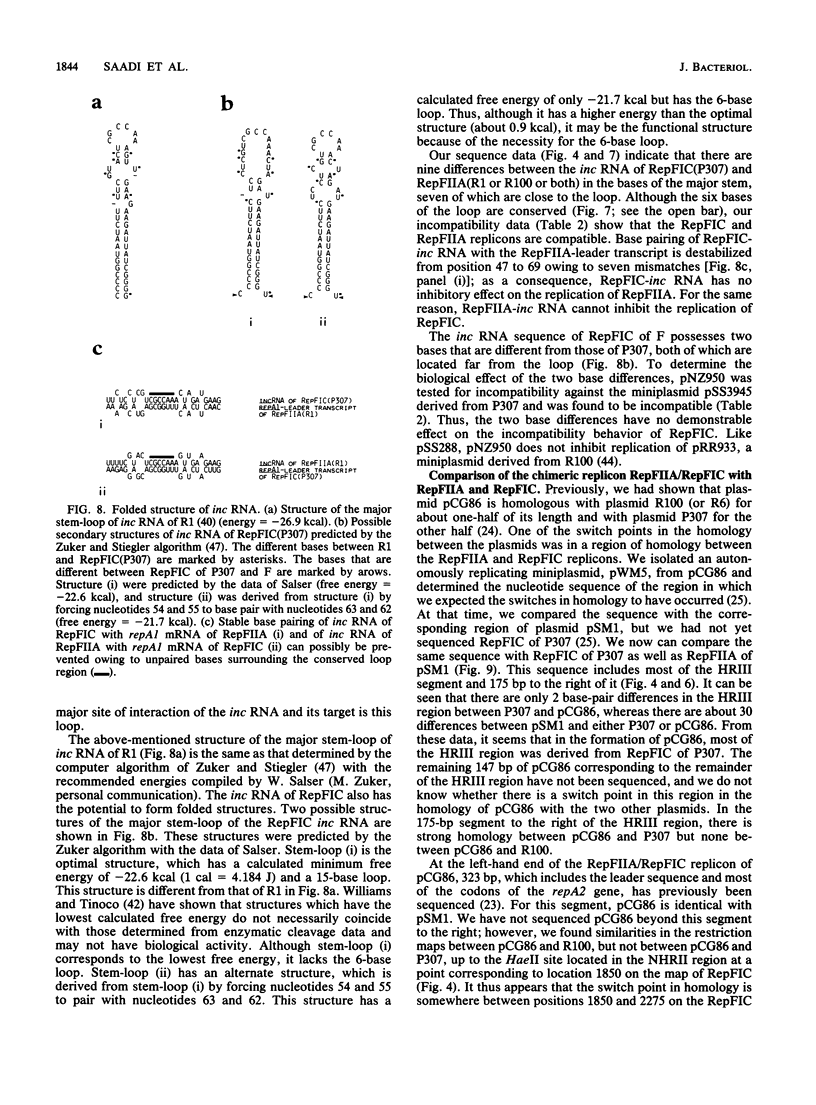
RepFIC is a basic replicon of IncFI plasmid P307 which is located within a 3.09-kilobase SmaI fragment. The nucleotide sequence of this region has been determined and shown to be homologous with the RepFIIA replicon of IncFII plasmids. The two replicons share three homologous regions, HRI, HRII, and HRIII, which are flanked by two nonhomologous regions, NHRI and NHRII. A comparison of coding regions reveals that the two replicons have several features in common. RepFIC, like RepFIIA, codes for a repA2 protein with its amino-terminal codons in HRI and its carboxy-terminal codons in NHRI. Although the codons for the repA1 proteins are located in NHRII, the DNA region containing a putative promoter, ribosomal binding site, and initiation codons is located in HRII. This region also codes for an inc RNA. There are nine base-pair differences between the inc RNA of RepFIIA and that of RepFIC, and as a result, RepFIC and RepFIIA replicons are compatible. An EcoRI fragment from the F plasmid which shows homology with RepFIC of P307 has also been sequenced. This fragment contains only a portion of RepFIC, including the genes for the putative repA2 protein and inc RNA. The region coding for a putative repA1 protein is interrupted by the transposon Tn1000 and shows no homology with the repA1 region of RepFIIA and RepFIC of P307. Our comparative and structural analyses suggest that RepFIC and RepFIIA, although different, have a similar replication mechanism and thus can be assigned to the same replicon family, which we designate the RepFIIA family.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Akimoto S., Ohnishi Y. R483 and F plasmid genes promoting RNA degradation: comparative restriction mapping. Microbiol Immunol. 1982;26(9):779–793. doi: 10.1111/j.1348-0421.1982.tb00224.x. [DOI] [PubMed] [Google Scholar]
- Andrés I., Slocombe P. M., Cabello F., Timmis J. K., Lurz R., Burkardt H. J., Timmis K. N. Plasmid replication functions. II. Cloning analysis of the repA replication region of antibiotic resistance plasmid R6-5. Mol Gen Genet. 1979 Jan 5;168(1):1–25. doi: 10.1007/BF00267929. [DOI] [PubMed] [Google Scholar]
- Bergquist P. L., Saadi S., Maas W. K. Distribution of basic replicons having homology with RepFIA, RepFIB, and RepFIC among IncF group plasmids. Plasmid. 1986 Jan;15(1):19–34. doi: 10.1016/0147-619x(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G., Frey J., Danbara H., Timmis K. N. Replication control mutations of plasmid R6-5 and their effects on interactions of the RNA-I control element with its target. J Bacteriol. 1983 Apr;154(1):429–436. doi: 10.1128/jb.154.1.429-436.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danbara H., Brady G., Timmis J. K., Timmis K. N. Regulation of DNA replication: "target" determinant of the replication control elements of plasmid R6-5 lies within a control element gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4699–4703. doi: 10.1073/pnas.78.8.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Womble D. D., Luckow V. A., Rownd R. H. Regulation of transcription of the repA1 gene in the replication control region of IncFII plasmid NR1 by gene dosage of the repA2 transcription repressor protein. J Bacteriol. 1985 Feb;161(2):544–551. doi: 10.1128/jb.161.2.544-551.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Gerhart E., Wagner H., Nordström K. Structural analysis of an RNA molecule involved in replication control of plasmid R1. Nucleic Acids Res. 1986 Mar 25;14(6):2523–2538. doi: 10.1093/nar/14.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givskov M., Molin S. Copy mutants of plasmid R1: effects of base pair substitutions in the copA gene on the replication control system. Mol Gen Genet. 1984;194(1-2):286–292. doi: 10.1007/BF00383529. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Lane D., Hill D., Caughey P., Gunn P. The mini-F primary origin. Sequence analysis and multiple activities. J Mol Biol. 1984 Dec 5;180(2):267–282. doi: 10.1016/s0022-2836(84)80004-2. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J., Molin S. The sites of action of the two copy number control functions of plasmid R1. Mol Gen Genet. 1982;187(3):486–493. doi: 10.1007/BF00332633. [DOI] [PubMed] [Google Scholar]
- Masai H., Kaziro Y., Arai K. Definition of oriR, the minimum DNA segment essential for initiation of R1 plasmid replication in vitro. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6814–6818. doi: 10.1073/pnas.80.22.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Nordström M., Nordström K. Control of replication of FII plasmids: comparison of the basic replicons and of the copB systems of plasmids R100 and R1. Plasmid. 1985 Mar;13(2):81–87. doi: 10.1016/0147-619x(85)90060-5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Vassino B., Ryder T., Ohtsubo E. A simple method for shortening a plasmid genome using a system of plasmid cointegration mediated by a Tn3 mutant. Gene. 1982 Dec;20(2):245–254. doi: 10.1016/0378-1119(82)90043-9. [DOI] [PubMed] [Google Scholar]
- Picken R. N., Mazaitis A. J., Maas W. K. High incidence of transposon Tn3 insertions into a replication control gene of the chimeric R/Ent plasmid pCG86 of Escherichia coli. J Bacteriol. 1984 Oct;160(1):430–433. doi: 10.1128/jb.160.1.430-433.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken R. N., Mazaitis A. J., Maas W. K. Restriction enzyme maps of the chimeric R/Ent plasmid pCG86 and the related ent plasmid P307. Plasmid. 1984 Jan;11(1):102–104. doi: 10.1016/0147-619x(84)90014-3. [DOI] [PubMed] [Google Scholar]
- Picken R. N., Mazaitis A. J., Saadi S., Maas W. K. Characterization of the basic replicons of the chimeric R/Ent plasmid pCG86 and the related Ent plasmid P307. Plasmid. 1984 Jul;12(1):10–18. doi: 10.1016/0147-619x(84)90062-3. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Young R. A., Steitz J. A., Grindley N. D., Guyer M. S. Transposition of the Escherichia coli insertion element gamma generates a five-base-pair repeat. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4882–4886. doi: 10.1073/pnas.76.10.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riise E., Molin S. Purification and characterization of the CopB replication control protein, and precise mapping of its target site in the R1 plasmid. Plasmid. 1986 May;15(3):163–171. doi: 10.1016/0147-619x(86)90034-x. [DOI] [PubMed] [Google Scholar]
- Robinson P., Bergquist P., Lane D. Analysis of a region in plasmid R386 containing two functional replicons. Plasmid. 1985 Jul;14(1):28–36. doi: 10.1016/0147-619x(85)90029-0. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Ohtsubo H., Ohtsubo E. Role of RNA transcripts in replication incompatibility and copy number control in antibiotic resistance plasmid derivatives. Nature. 1981 Apr 30;290(5809):794–797. doi: 10.1038/290794a0. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- Saadi S., Maas W. K., Bergquist P. L. RepFIC, a basic replicon of IncFI plasmids that has homology with a basic replicon of IncFII plasmids. Plasmid. 1984 Jul;12(1):61–64. doi: 10.1016/0147-619x(84)90067-2. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D., Yaish S., Archer J., Sherratt D. Multimer resolution systems of ColE1 and ColK: localisation of the crossover site. Mol Gen Genet. 1985;201(2):334–338. doi: 10.1007/BF00425680. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Andrés I., Slocombe P. M. Plasmid incompatibility: cloning analysis of an incFII determinant of R6-5. Nature. 1978 May 4;273(5657):27–32. doi: 10.1038/273027a0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Palchaudhuri S. Incompatibility repressor in a RepA-like replicon of the IncFI plasmid ColV2-K94. J Bacteriol. 1986 Jun;166(3):1106–1112. doi: 10.1128/jb.166.3.1106-1112.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. L., Jr, Tinoco I., Jr A dynamic programming algorithm for finding alternative RNA secondary structures. Nucleic Acids Res. 1986 Jan 10;14(1):299–315. doi: 10.1093/nar/14.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Luckow V. A., Wu R. P., Rownd R. H. Analysis of the individual regulatory components of the IncFII plasmid replication control system. J Bacteriol. 1985 Feb;161(2):534–543. doi: 10.1128/jb.161.2.534-543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Dong X., Wu R. P., Luckow V. A., Martinez A. F., Rownd R. H. IncFII plasmid incompatibility product and its target are both RNA transcripts. J Bacteriol. 1984 Oct;160(1):28–35. doi: 10.1128/jb.160.1.28-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Sampathkumar P., Easton A. M., Luckow V. A., Rownd R. H. Transcription of the replication control region of the IncFII R-plasmid NR1 in vitro and in vivo. J Mol Biol. 1985 Feb 5;181(3):395–410. doi: 10.1016/0022-2836(85)90228-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]




