Abstract
The nucleotide sequence of the streptothricin acetyltransferase (STAT) gene from streptothricin-producing Streptomyces lavendulae predicts a 189-amino-acid protein of molecular weight 20,000, which is consistent with that determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the purified enzyme. The amino acid composition and the NH2-terminal sequence determined by using the purified protein are in good agreement with those predicted from the nucleotide sequence, except for the absence of the NH2-terminal methionine in the mature protein. High-resolution S1 nuclease protection mapping suggests that transcription initiates at or near the adenine residue which is the first position of the translational initiation triplet (AUG) of STAT. Another open reading frame located just upstream of the STAT gene was detected and contains a region bearing a strong resemblance to DNA-binding domains which are conserved in known DNA-binding proteins. By addition of promoter signals and a synthetic ribosome-binding (Shine-Dalgarno) sequence at an appropriate position upstream of the STAT translational start codon, the STAT gene confers streptothricin resistance on Escherichia coli and Bacillus subtilis. The STAT coding sequence with both the promoter of a B. subtilis cellulase gene and a synthetic Shine-Dalgarno sequence was functionally expressed in Streptomyces lividans, which suggests that the addition of an artificial leader upstream of the translational initiation codon (AUG) does not significantly influence the translation of STAT.
Full text
PDF


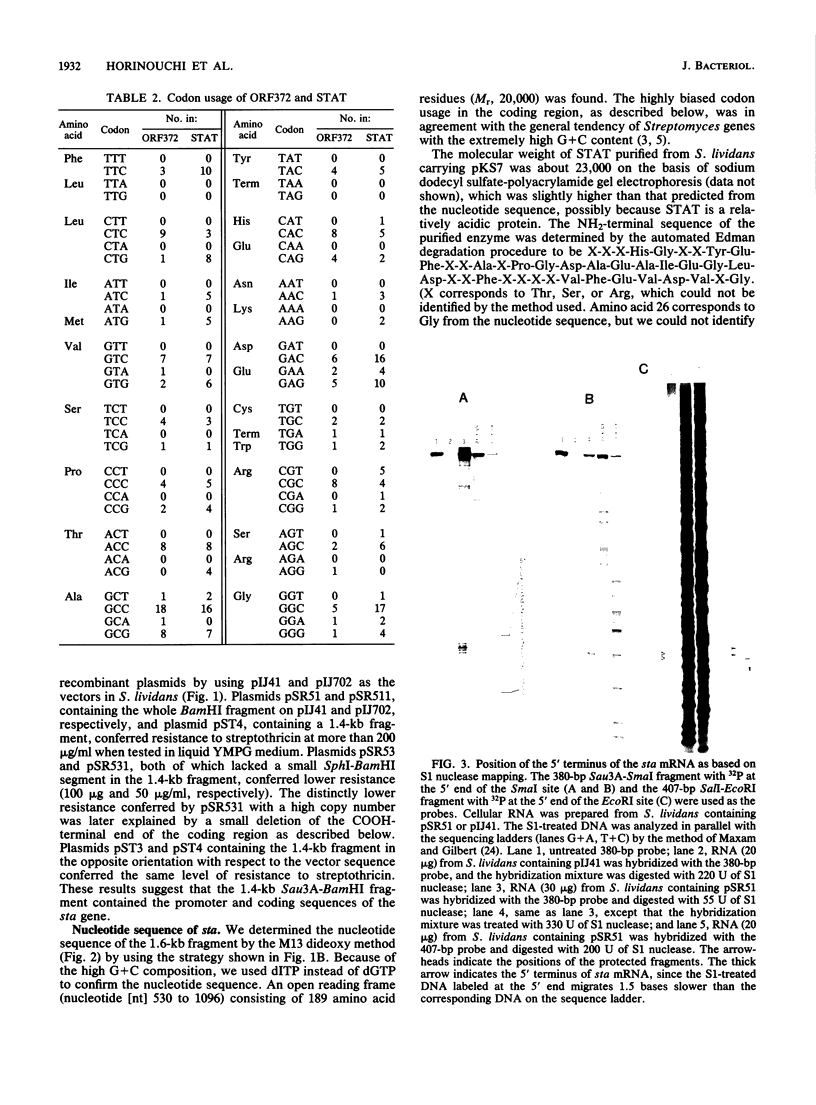

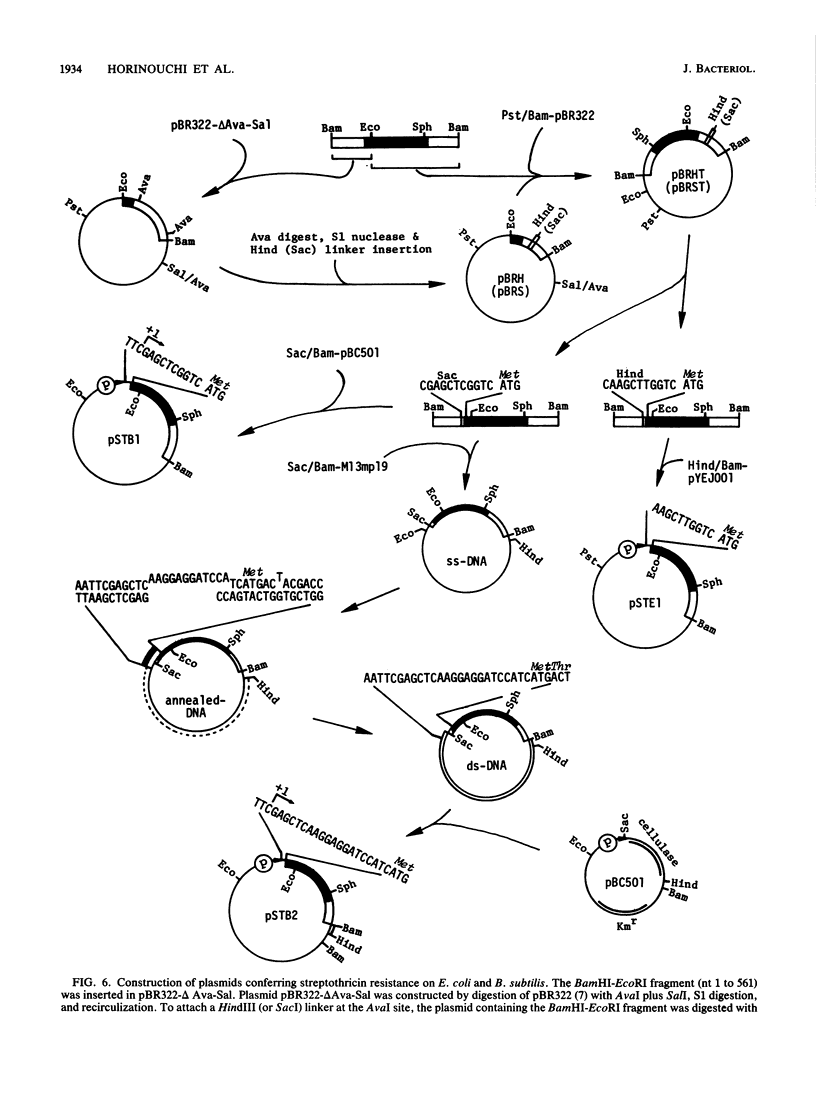
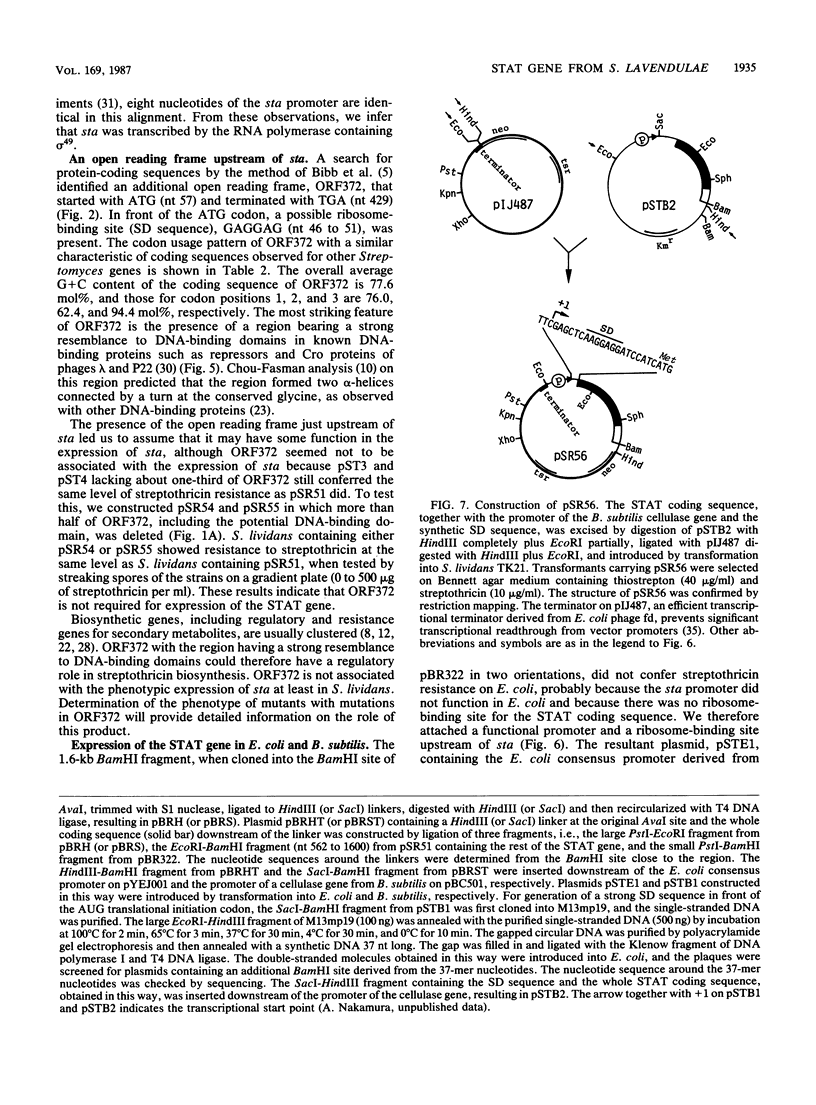


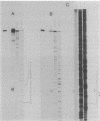
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernan V., Filpula D., Herber W., Bibb M., Katz E. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene. 1985;37(1-3):101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 1985 Jul;4(7):1893–1897. doi: 10.1002/j.1460-2075.1985.tb03866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T., Thompson C. J. Gene cloning in Streptomyces. Curr Top Microbiol Immunol. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Feitelson J. S., Malpartida F., Hopwood D. A. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2). J Gen Microbiol. 1985 Sep;131(9):2431–2441. doi: 10.1099/00221287-131-9-2431. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Kieser T., Wright H. M., Bibb M. J. Plasmids, recombination and chromosome mapping in Streptomyces lividans 66. J Gen Microbiol. 1983 Jul;129(7):2257–2269. doi: 10.1099/00221287-129-7-2257. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Beppu T. Construction and application of a promoter-probe plasmid that allows chromogenic identification in Streptomyces lividans. J Bacteriol. 1985 Apr;162(1):406–412. doi: 10.1128/jb.162.1.406-412.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Suzuki H., Beppu T. Nucleotide sequence of afsB, a pleiotropic gene involved in secondary metabolism in Streptomyces coelicolor A3(2) and "Streptomyces lividans". J Bacteriol. 1986 Oct;168(1):257–269. doi: 10.1128/jb.168.1.257-269.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Uozumi T., Beppu T. Cloning and characterization of the streptothricin-resistance gene which encodes streptothricin acetyltransferase from Streptomyces lavendulae. J Antibiot (Tokyo) 1986 May;39(5):688–693. doi: 10.7164/antibiotics.39.688. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. 1984 May 31-Jun 6Nature. 309(5967):462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y. Cro repressor protein and its interaction with DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):427–433. doi: 10.1101/sqb.1983.047.01.050. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Ohnuki T., Imanaka T., Aiba S. Self-cloning in Streptomyces griseus of an str gene cluster for streptomycin biosynthesis and streptomycin resistance. J Bacteriol. 1985 Oct;164(1):85–94. doi: 10.1128/jb.164.1.85-94.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Tatti K. M., Moran C. P., Jr Promoter recognition by sigma-37 RNA polymerase from Bacillus subtilis. J Mol Biol. 1984 May 25;175(3):285–297. doi: 10.1016/0022-2836(84)90349-8. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Uchiyama H., Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene. 1985;38(1-3):103–110. doi: 10.1016/0378-1119(85)90208-2. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Zalacain M., González A., Guerrero M. C., Mattaliano R. J., Malpartida F., Jiménez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986 Feb 25;14(4):1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]