Abstract
The effects of two peptidyl-transferase inhibitors, anisomycin and sparsomycin, on ribosomal frameshifting efficiencies and the propagation of yeast double-stranded RNA viruses were examined. At sublethal doses in yeast cells these drugs specifically alter the efficiency of −1, but not of +1, ribosomal frameshifting. These compounds promote loss of the yeast L-A double-stranded RNA virus, which uses a programmed −1 ribosomal frameshift to produce its Gag-Pol fusion protein. Both of these drugs also change the efficiency of −1 ribosomal frameshifting in yeast and mammalian in vitro translation systems, suggesting that they may have applications to control the propagation of viruses of higher eukaryotes, which also use this translational regulatory mechanism. Our results offer a new set of antiviral agents that may potentially have a broad range of applications in the clinical, veterinary, and agricultural fields.
Programmed −1 ribosomal frameshifting is a mode of regulating gene expression used predominantly by RNA viruses and a subset of bacterial genes to induce elongating ribosomes to shift reading frame in response to specific mRNA signals (reviewed in refs. 1–3). In the yeast L-A double-stranded RNA (dsRNA) virus, a −1 ribosomal frameshift event is responsible for the production of a Gag-Pol fusion protein (4). M1, a satellite dsRNA virus of L-A that encodes a secreted killer toxin, is encapsulated and replicated using the Gag and Gag-Pol gene products synthesized by the L-A virus (reviewed in ref. 5). Maintaining the appropriate ratio of Gag to Gag-Pol is critical for maintenance of the M1 virus (6). A screen for mutations that increased the programmed −1 ribosomal frameshift efficiencies identified nine chromosomal mof mutants (maintenance of frame), many of which promoted the loss of the satellite killer M1 (6–8).
One of the most extensively studied examples of the link between the processes of mRNA translation and turnover is the observation that premature translational termination can enhance decay rates of mRNAs in a process called nonsense-mediated mRNA decay (9). In the yeast Saccharomyces cerevisiae, mutations in the UPF1, UPF2, and UPF3 genes inactivate the nonsense-mediated mRNA decay pathway and promote suppression of certain nonsense alleles (10–12). Recent results also have demonstrated that the Upf1 protein (Upf1p) is a modulator of nonsense-mediated mRNA decay, translation termination, and programmed −1 frameshifting (13–15). Molecular and genetic analysis of the mof4–1 allele demonstrated that it is allelic to UPF1 (15). Further, mof4–1 strains are more sensitive to the aminoglycoside antibiotic paromomycin and the efficiency of programmed −1 ribosomal frameshifting increased in a mof4–1 strain grown in the presence of this drug (15). These observations support the hypothesis that certain classes of translational inhibitors may specifically affect programmed −1 ribosomal frameshifting and promote virus loss in wild-type cells.
Many viruses of clinical, veterinary, and agricultural importance use programmed frameshifting for the production of their structural and enzymatic gene products (reviewed in ref. 16), and we suggest that programmed ribosomal frameshifting is a unique target to identify antiviral agents. Because both −1 and +1 ribosomal frameshifting are driven by ribosomal pause events (reviewed in refs. 1 and 2), it is reasonable to assume that changing the kinetic parameters of these pauses may, in turn, alter frameshift efficiencies. Further, because frameshifting in either direction occurs during translation elongation, the kinetics of ribosomal pausing are subject to the three kinetically rate limiting steps in this cycle: (i) selection and insertion of cognate aminoacyl-tRNA (aa-tRNA) into the ribosomal A-site, (ii) peptidyl transfer, and (iii) translocation. Programmed −1 ribosomal frameshifting occurs while both ribosomal A- and P-sites are occupied by cognate tRNAs, i.e. after step 1 and before step 3. In contrast, Ty1-promoted +1 ribosomal frameshifting occurs while only the P-site is occupied by peptidyl-tRNA, i.e. after step 3 and before step 1. Thus, these two mechanisms should be experimentally separable. We have developed in vivo and in vitro programmed frameshifting assays to monitor the effects of drugs that are known to affect the translation elongation process to determine how they affect programmed ribosomal frameshifting. Here we report that peptidyl-transferase inhibitors specifically affect the efficiency of programmed ribosomal frameshifting in the −1 direction and that these agents also promote loss of the yeast dsRNA killer viruses.
MATERIALS AND METHODS
Strains and Media.
S. cerevisiae strains used in this study were JD88 [MATa ura3–52 lys2–801 ade2–10 trp1-Δ1 (L-AHNB) (M1)], JD96 (which is JD88 cured of the killer viruses), and 5×47 (killer test strain (MATa/MATα his1/+ trp1/+ ura3/+ K− R−). YPAD, complete synthetic medium, and 4.7 methylene blue plates for testing the killer phenotype were as previously reported (7). Anisomycin was purchased from Sigma. Sparsomycin was generously provided by S. Pestka (Robert Wood Johnson Medical School).
Genetic Methods.
Transformation of yeast and Escherichia coli was performed as described previously (4, 15). Killer tests and β-galactosidase (β-gal) assays were as described previously (4, 6, 7, 15). Measurement of the effects of anisomycin and sparsomycin on ribosomal frameshifting efficiencies involved inoculating selective medium containing the indicated concentrations of drugs with cells harboring the ribosomal frameshift reporter plasmids to an OD595 = 0.1. Cultures were subsequently incubated for 5 hr at 30°C, after which β-gal activities and ribosomal frameshifting efficiencies were determined by calculating the ratios of β-gal activity in cells harboring the frameshift test vectors (−1 = pF8; +1 = pJD104) to β-gal activity in cells harboring the 0-frame control (pTI25) and multiplying by 100% (4, 17). All assays were performed in triplicate. To measure rates of killer phenotype loss, JD88 cells were inoculated into YPAD containing the indicated concentrations of either sparsomycin or anisomycin and incubated at 30°C for 1 to 5 days, aliquots of cells were removed every 24 hr, streaked for single colonies onto YPAD, and subsequently replica-plated to 4.7 methylene blue killer indicator plates. The frequency of killer loss was measured by the ratio of Killer−/total colonies. At least 100 individual colonies were assayed for each drug concentration and each time point.
Plasmids.
The ribosomal frameshift assay plasmids pTI25 (0-frame control), pF8, and pJD104 (−1 and +1 ribosomal frameshift test plasmids) were used to determine the efficiency of L-A-promoted −1, and Ty1-promoted +1 ribosomal frameshifting respectively, and are as described previously (refs. 4 and 17; see Fig. 1A). Plasmids p315-JD85-ter and p315-JD86-ter (15) are −1 ribosomal frameshift and 0-frame reporter plasmids, respectively, which were used for analysis of lacZ mRNA abundance.
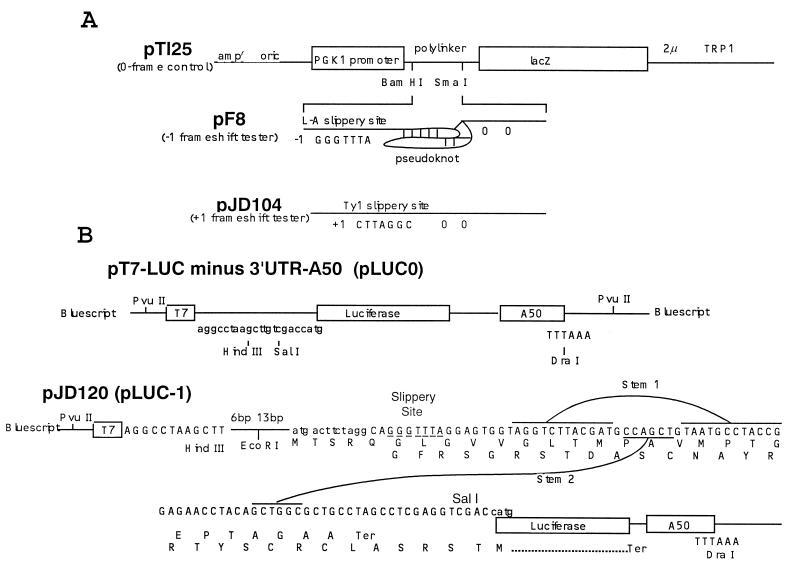
Figure 1.
(A) Vectors used to measure ribosomal frameshift efficiencies in vivo. Plasmid pTI25 and pF8 are as described previously (4), and plasmid pJD104 is as described previously (17). Transcription is driven from the PGK1 promoter and uses the PGK1 translation initiation codon. In pTI25, the bacterial lacZ gene is in the 0-frame with respect to the start site. In plasmid pF8 the lacZ gene is positioned 3′ of the L-A virus frameshift site and in the −1 frame relative to the translation start site. Plasmid pJD104 is derived from pF8 (17) and contains the Ty1 +1 ribosomal frameshift signal 5′ of the lacZ gene. The lacZ gene is inserted in the +1 translation reading frame relative the start of translation. Termination codons in the −1 and +1 frames (pF8 and pJD104, respectively) are located 5′ of the frameshift signals and two 0-frame termination codons are 3′ of the frameshift signal. (B) Vectors to measure ribosomal frameshift efficiencies in vitro. pT7-LUC minus 3′UTR-A50 (LUC0) (18) is the 0-frame reference plasmid. pJD120 (LUC-1) contains the L-A −1 ribosomal frameshift signal cloned into the HindIII/SalI sites of pLUC0. The luciferase coding sequence is in the −1 reading frame with respect to the translational start site such that all luciferase activity must result as a consequence of a −1 ribosomal frameshift event. pLUC0 and pLUC-1 were linearized with DraI and synthetic 7-methyl-Gppp-capped, poly(A)+-tailed transcripts were made using T7 RNA polymerase.
Plasmid pT7-LUC minus 3′UTR-A50 (18) (referred to here as pLUC0; see Fig. 1B) was used to produce synthetic 0-frame luciferase-encoding mRNAs. The vector for the production of the −1 ribosomal frameshift luciferase reporter mRNA was constructed as follows: Single-stranded, uracil+ DNA from pJD90 [which contains the minimal L-A −1 ribosomal frameshift signal derived from pF′8–5D17/3D32XS (4) cloned into BlueScript SK−] was first mutagenized in vitro with the synthetic oligonucleotide 5′-CTGGCGCTGCCTAGCTTCGAGGTCGACGATCCACTAGTTC-3′ to create a unique SalI site (bold); next, the resulting plasmid (pJD119) was digested with HindIII and SalI, and the 123-bp dsDNA fragment was cloned into HindIII/SalI-digested pLUC0 to construct the −1 ribosomal frameshift luciferase reporter pJD120 (pLUC-1, Fig. 1B). In this plasmid, the luciferase gene is 3′ of the L-A −1 ribosomal frameshift signal and in the −1 frame with respect to the AUG translational start site.
Nucleic Acid Analyses.
dsRNA of L-A and M1 viruses was prepared as described (19), separated by electrophoresis through 1.2% agarose gels, denatured in the gels in two changes of 30 min each of 50% formamide, 9.25% formaldehyde, 1× Tris-acetate-EDTA at room temperature and transferred to nitrocellulose in 20× standard saline citrate. L-A and M1 (+) strand RNA probes were labeled with [α-32P]UTP, hybridized to blots, and washed as described in ref. 7. The abundances of lacZ reporter mRNA (from p315-JD85-ter), the CYH2 precursor, and CYH2 mRNAs were determined as described previously (15).
In Vitro Translation.
pLUC0 and pLUC-1 were linearized with DraI and synthetic 7-methyl-Gppp-capped, poly(A)+- tailed mRNAs were made using T7 RNA polymerase and a MessageMachine kit (Ambion). In vitro translations used 20 ng of either LUC0 or LUC-1 mRNA and were performed in triplicate at the indicated concentrations of either anisomycin or sparsomycin at 25°C. The Reticulocyte Lysate IVT kit (Ambion) was used for reticulocyte in vitro translation assays. JD96 cells were used as the source of translation-competent yeast extracts, which were prepared by the method of ref. 20 and modified according to ref. 21. Reactions were incubated for 1 hr, stopped on dry ice, and thawed on ice, and luciferase activities were determined using a Turner 20/20 luminometer.
RESULTS
Assays to Monitor the Effect of Compounds on Programmed Ribosomal Frameshifting.
The programmed ribosomal frameshifting assays described below are the basis for our initial screens for compounds that affect these processes. Our approach is to search for compounds that alter programmed ribosomal frameshifting efficiencies and promote loss of the M1 virus. Because programmed −1 ribosomal frameshifting events occur while both the ribosomal A- and P-sites are occupied by cognate tRNAs during translation elongation, we postulated that compounds that affect the peptidyl-transferase reaction may affect this process.
Methods to measure efficiencies of programmed ribosomal frameshifting in vivo have been previously described (see Fig. 1A; refs. 4, 6–8, 11, 17). A series of reporter plasmids are used in which transcription is driven from the yeast PGK1 promoter and terminates at the PGK1 polyadenylylation site (15). A translational start codon is followed by a multiple cloning site, followed by the E. coli lacZ gene. Plasmid pTI25 serves as the 0-frame control in that lacZ is in the 0-frame with respect to the translational start site (4). In pF8, an L-A-derived programmed −1 ribosomal frameshift signal is cloned into the polylinker of pTI25, and the lacZ gene is in the −1 frame with respect to the translational start site. In this construct, the protein coding sequence from the lacZ gene will be translated only if the ribosome shifts frame in the −1 direction (Fig. 1A). The Ty1 retrotransposable element of yeast uses a programmed +1 ribosomal frameshift to produce its Gag-Pol fusion protein (22), a process that occurs independently of the peptidyl-transfer reaction. Thus as a control, the +1 frameshift reporter plasmid (pJD104, Fig. 1A; ref. 17) contains the lacZ gene inserted 3′ of the Ty1 frameshift sequence and in the +1 frame relative to the start of translation. In pJD104, the protein coding sequence from the lacZ gene will be translated only if the ribosome shifts frame in the +1 direction (Fig. 1A). The efficiency of −1 ribosomal frameshifting is calculated by determining the ratio of β-gal activities measured in cells harboring the −1 frameshift reporter plasmid pF8 to those harboring the 0-frame control plasmid pTI25. Similarly, calculation of +1 ribosomal frameshifting efficiency is based on the pJD104 to pTI25 β-gal ratios. Because the efficiency of programmed ribosomal frameshifting is based on the ratio of frameshift reporter to 0-frame control β-gal activities, the efficiencies of programmed ribosomal frameshifting were always normalized to the effects that the drugs have on translation by monitoring β-gal activity of the zero-frame control in cells grown in the presence of the indicated drug. The fold change in programmed frameshifting was determined by calculating the ratio of frameshifting efficiency in the presence of the drugs versus the frameshifting efficiency in cells grown in the absence of any drug. The presence of the M1 dsRNA virus can be monitored by replica plating colonies on a lawn of cells that are sensitive to the killer toxin. Cells maintaining the M1 virus secrete the killer toxin, creating a ring of growth inhibition (6).
Anisomycin and Sparsomycin Alter the Efficiency of −1, but Not +1, Ribosomal Frameshifting in Vivo.
A preliminary screen demonstrated that anisomycin and sparsomycin, two compounds that were previously identified to affect steps in the peptidyl transfer center (reviewed in ref. 23), affected programmed ribosomal frameshifting (see below), whereas general inhibitors of translation such as paromomycin, cycloheximide, and hygromycin had no such effects (ref. 11 and data not shown). To determine a suitable range of concentrations to analyze the effects of these drugs on programmed frameshifting, cell growth was monitored by measuring the doubling time of cells grown in rich medium at various drug concentrations. Anisomycin concentrations ranging from 0.76 to 3.80 μM, and sparsomycin concentrations ranging from 0.52 to 2.60 μM inhibited overall cellular growth rates by less than 30% (data not shown). These concentrations of the drugs were used to investigate their in vivo effects on programmed frameshifting.
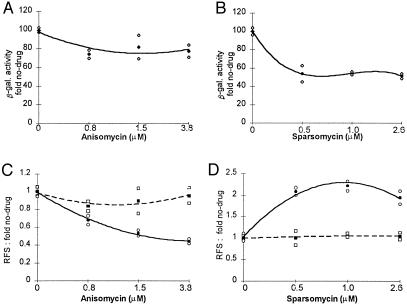
The effect of these drugs on programmed −1 and +1 ribosomal frameshifting efficiencies in vivo were monitored in selective medium containing either no drug or various concentrations of anisomycin or sparsomycin. Cells containing either the frameshift or zero-frame reporter constructs were grown at 30°C in the presence or absence of the indicated drugs for 5 hr (Fig. 2). The β-gal activities in these cells then were monitored. β-gal activity in cells harboring the 0-frame control plasmid pTI25 decreased less than 20% when grown at the highest concentration of anisomycin tested and less than 50% in the presence of the highest concentrations of sparsomycin analyzed. The fold changes in programmed −1 frameshifting as a consequence of growing the cells in the presence of the drugs were determined, and the results are shown in Fig. 2. Within these ranges of drug concentrations, the efficiency of −1 ribosomal frameshifting decreased in the presence of anisomycin, whereas, conversely, sparsomycin increased −1 ribosomal frameshifting efficiencies (Fig. 2 C and D). These drugs were specific to −1 programmed frameshifting because neither drug had any effect on +1 ribosomal frameshifting (Fig. 2 C and D).
Figure 2.
Anisomycin and sparsomycin specifically affect −1 ribosomal frameshifting efficiencies in vivo. JD88 cells containing the frameshift indicator plasmids pTI25, pF8, or pJD104 were cultured in selective medium in the presence of the indicated antibiotic concentrations for 5 hr, after which β-gal activities were determined. The filled symbol represents the average of at least three independent experiments. Open symbols represent high and low margins of error. (A and B) β-gal activities produced from pTI25 as a percentage of the no-drug controls. (C and D) The fold changes in −1 or +1 ribosomal frameshifting efficiencies (RFS) as compared with the no-drug controls [−1 (solid line) = 1.8%; +1 (dashed line) = 5.5%].
Anisomycin and Sparsomycin Do Not Affect the Abundance of the Reporter Transcripts.
The apparent changes in ribosomal frameshifting efficiencies could result from changes in the abundance of the −1 ribosomal frameshift lacZ mRNA, which the cell may see as a nonsense-containing mRNA (see ref. 15). To address this possibility, RNAs were extracted from mid-log phase cells grown in different concentrations of anisomycin or sparsomycin, and the abundance of the lacZ frameshift reporter mRNA was determined by Northern blotting analysis. As a loading control, the blots also were probed for the U3 snRNA and CYH2 transcript and precursor. Probing for CYH2 provides an independent monitor of the effects of these drugs on the abundance of another RNA that is a substrate for nonsense-mediated mRNA decay, because the inefficiently spliced endogenous CYH2-precursor mRNA is normally rapidly degraded but is stabilized in strains in which the nonsense-mediated mRNA decay pathway is inactivated (9). Quantitation of the hybridizing bands revealed that the abundances of the lacZ frameshift reporter mRNA, normalized to the U3 snRNA, were equivalent at all concentrations of drugs tested, and were comparable to the abundances observed in cells not treated with the drugs (Table 1). Further, these drugs did not affect the abundance of the CYH2 precursor (data not shown). These results indicated that drug-induced changes in programmed −1 frameshifting were most likely a consequence of affecting −1 ribosomal frameshifting efficiencies and were not a consequence of altering the stability of these mRNAs.
Table 1.
Effect of drug addition on the abundance of the reporter transcript
| Antibiotic | Conc., μM | mRNA abundance* |
|---|---|---|
| None | 1.0 | |
| Anisomycin | 0.76 | 0.854 |
| 1.50 | 0.908 | |
| 3.80 | 0.937 | |
| 7.60 | 0.937 | |
| Sparsomycin | 0.52 | 0.9 |
| 1.00 | 1.0 | |
| 2.60 | 0.96 | |
| 5.20 | 0.98 |
The abundances of the reporter transcript in the presence of different concentration of the antibiotics, normalized to the abundance of the U3 snRNA, are expressed as the ratio of the no-drug control, which is arbitrarily taken as 1.0.
Anisomycin and Sparsomycin Promote Virus Loss.
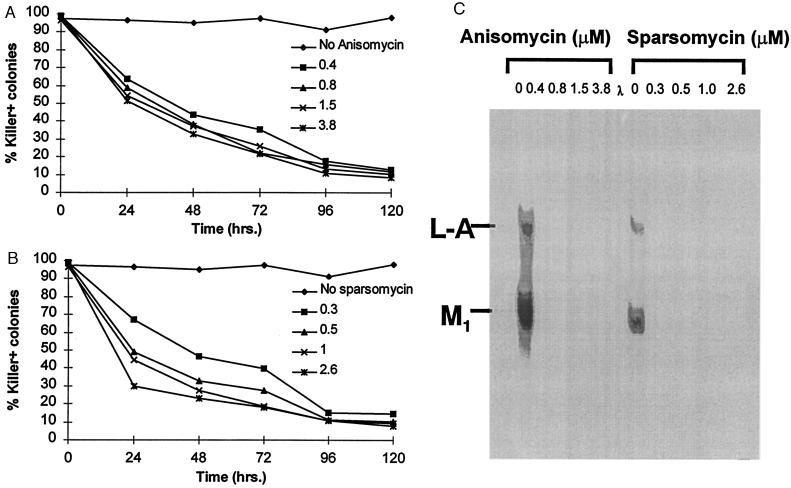
Changing the efficiency of −1 ribosomal frameshifting alters the ratio of Gag to Gag-Pol proteins available for viral particle assembly, consequently interfering with viral propagation (6, 7). Therefore, we asked whether sparsomycin and anisomycin promoted loss of L-A and/or of its satellite virus, M1. Wild-type cells were cultured in medium containing different concentrations of either anisomycin or sparsomycin for up to 5 days, and every 24 hr aliquots of cells were streaked for single colonies onto rich medium. The resulting colonies were replica-plated onto a lawn of cells sensitive to the killer toxin. Cells maintaining the M1 virus secrete the killer toxin, creating a ring of growth inhibition, whereas cells that have lost M1 do not (6). The results of this assay demonstrated that both drugs promoted rapid loss of the killer phenotype (Fig. 3 A and B). Virus loss correlated best with increased time of incubation with the drugs (Fig. 3 A and B), suggesting a dilution of the original pool of intracellular virus particles.
Figure 3.
Anisomycin and sparsomycin promote virus loss. JD88 cells were grown in rich medium in the presence of the indicated concentrations of drugs for 1–5 days, and aliquots of cells were removed every 24 hr, streaked for single colonies onto rich medium lacking drugs, and replicated to killer indicator plates. (A and B) Rates of killer loss were calculated as described in Materials and Methods. Anisomycin and sparsomycin concentrations (μM) are indicated. (C) Total nucleic acids were extracted from control (Killer+) and non-Killer colonies, separated by agarose gel electrophoresis, transferred to nitrocellulose and hybridized with L-A and M1 (+) strand probes, and visualized by autoradiography as described in Materials and Methods.
To determine whether loss of the killer phenotype was a consequence of loss of virus and not interference with translation and processing of the killer toxin, a non-Killer (K−) colony from each drug concentration in the 72-hr data set was picked at random, total nucleic acids were extracted (19), and equal amounts of nucleic acids were separated through a nondenaturing agarose gel. The RNAs were transferred to nitrocellulose and hybridized with [32P]CTP labeled L-A and M1 (+) strand RNA probes. The results demonstrated that these samples do not hybridize with either of the probes, confirming that loss of the killer phenotype was a consequence of loss of L-A and M1 (Fig. 3C). These data support the hypothesis that drug induced changes in the efficiency of −1 ribosomal frameshifting interfered with the assembly and propagation of L-A viral particles.
Anisomycin and Sparsomycin Affect the Efficiency of −1 Ribosomal Frameshifting in Two Different in Vitro Translation Systems.
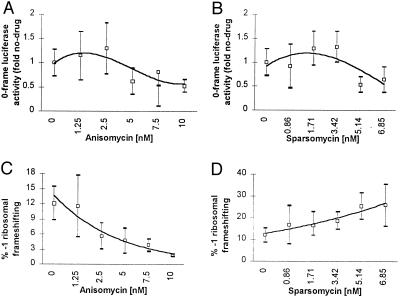
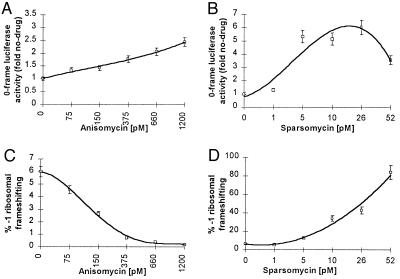
The effects of anisomycin and sparsomycin on programmed −1 frameshifting were monitored in a yeast in vitro translation system using a luciferase reporter system. The system used translation competent yeast extracts from a strain lacking the killer virus (L-A-o, L-BC-o wild-type strain) (21). 7-methyl-G-capped and polyadenylylated transcripts containing the luciferase protein coding region either lacking (0-frame control, LUC0) or containing (LUC-1) a −1 ribosomal frameshift site were prepared in vitro (see Fig. 1B). These mRNAs were added to the translation extract, and the amount of luciferase synthesized was determined. The efficiency of programmed frameshifting was determined by multiplying the ratio of LUC-1/LUC0 luciferase activities by 100%. The results of these experiments are shown in Fig. 4 and demonstrate that the effects of these drugs on programmed −1 frameshifting observed in vivo were reproducible in this in vitro translation system (compare Figs. 4C with 2C and 4D with 2D). Increasing concentrations of anisomycin decreased the efficiency of −1 ribosomal frameshifting, whereas increasing concentrations of sparsomycin increased −1 ribosomal frameshifting efficiencies. As determined by monitoring luciferase activities in the samples containing the zero-frame control reporter construct, approximately 40% inhibition of overall translation was observed at the highest concentrations used (Fig. 4 A and B).
Figure 4.
Anisomycin and sparsomycin affect −1 ribosomal frameshifting efficiencies in a yeast extract based in vitro translation system. Synthetic 7-methyl-Gppp-capped, polyadenylylated mRNAs were transcribed from DraI-digested pLUC0 and pLUC-1, and 20 ng of LUC0 or LUC-1 mRNA was used for each in vitro translation reaction in the presence of the indicated concentrations of anisomycin or sparsomycin. The open squares represent the average of at least three independent experiments. High and low margins of error are indicated. A (anisomycin) and B (sparsomycin): Changes in 0-frame luciferase as a percentage of no-drug controls. C (anisomycin) and D (sparsomycin): % −1 ribosomal frameshifting.
We also tested whether anisomycin and sparsomycin affect the efficiency of −1 ribosomal frameshifting in mammalian translation extracts. The same transcripts described above (LUC0 and LUC-1 mRNAs) were added to a rabbit reticulocyte translation system in the presence of various concentrations of both drugs and the effect on translation and programmed −1 ribosomal frameshifting efficiencies were monitored as described above. The reticulocyte extracts were approximately 10 times more sensitive to anisomycin and 100 times more sensitive to sparsomycin than were the yeast extracts. In this range of concentrations, both drugs stimulated overall production of luciferase from the LUC0 mRNA (Fig. 5A and B). Fig. 5 C and D demonstrates that, with regard to their effects on −1 ribosomal frameshifting efficiencies, the same general trends seen in the yeast based in vitro translation system were recapitulated in the rabbit reticulocyte system. Generally, anisomycin inhibited programmed frameshifting, whereas sparsomycin stimulated programmed −1 ribosomal frameshifting. Specifically, the effects of the drugs were more pronounced in the reticulocyte system. The highest concentration of anisomycin (12 nM) decreased −1 ribosomal frameshifting efficiencies to only 3% of the no drug control, effectively shutting it down (Fig. 5C). The highest concentration of sparsomycin stimulated −1 ribosomal frameshifting 14-fold (Fig. 5D).
Figure 5.
Anisomycin and sparsomycin affect −1 ribosomal frameshifting efficiencies in a rabbit reticulocyte in vitro translation system. Synthetic 7-methyl-Gppp-capped, polyadenylylated mRNAs were transcribed from DraI-digested pLUC0 and pLUC-1, and 20 ng of LUC0 or LUC-1 mRNA were used for each in vitro translation reaction in the presence of the indicated concentrations of anisomycin or sparsomycin. The open squares represent the average of at least three independent experiments. High and low margins of error are indicated. A (anisomycin) and B (sparsomycin): Changes in 0-frame luciferase as the fold of no-drug controls. C (anisomycin) and D (sparsomycin): % −1 ribosomal frameshifting.
DISCUSSION
We previously have described a family of mutant genes in yeast (mof mutants) that increased the efficiency of −1 ribosomal frameshifting as promoted by the L-A dsRNA virus frameshift signal (6–8), and identified mof4–1 as a mutant allele of the yeast UPF1 gene that affects both programmed −1 frameshifting and nonsense-mediated mRNA decay (15). Strains harboring the mof4–1 allele demonstrated greater sensitivity to paromomycin, a drug known to enhance translational misreading, and increased the efficiency of programmed −1 ribosomal frameshifting in a mof4–1 strain. Based on these findings and on the current models describing programmed ribosomal frameshifting, we speculated that certain compounds known to affect the ribosomal peptidyl transferase center may be able to recapitulate the effects of these mutations, resulting in loss of the killer virus. We hypothesized that, at subtoxic dosages, such antibiotics still may have an effect on how the aa-tRNAs and other factors interact with the ribosome, leading to changes in programmed −1 frameshifting. Further, because Ty1-promoted +1 ribosomal frameshifting occurs when the ribosomal A-site is vacant and the P-site is occupied by peptidyl-tRNA (22), this process should not be affected by peptidyl-transferase inhibitors. This hypothesis proved correct, because sparsomycin and anisomycin, two compounds known to interact with the peptidyl transfer center of a ribosome (reviewed in ref. 23), altered programmed −1 ribosomal frameshifting efficiencies in vivo but had no effect on +1 frameshifting (Fig. 2), and promoted loss of both L-A and M1 (Fig. 3C). Neither sparsomycin nor anisomycin affected the stability of the nonsense containing lacZ transcript, suggesting that these compounds affected the translation apparatus, perhaps by altering the likelihood of the A- and P-site aa- and peptidyl-tRNAs to slip.
Interestingly, sparsomycin and anisomycin affected programmed frameshifting in different ways (Figs. 2, 4, and 5). The known actions of these drugs may explain these different effects. Anisomycin preferentially inhibits binding of the aa-tRNA stem to the acceptor site of the peptidyl-transferase center (24). Thus, anisomycin, destabilizes the A-site specific tRNA–ribosome interaction at the level of the acceptor stem of the aa-tRNA. A −1 frameshift further destabilizes the the A-site specific tRNA–ribosome interaction at the level of the codon/anticodon base pairing. The sum of these two suboptimal interactions of the A-site aa-tRNA with the rest of the translational complex may activate the translational proofreading apparatus, such that translation is aborted preferentially in frameshifted ribosomes. Thus, anisomycin would effectively select against frameshifted ribosomes. Sparsomycin stimulates binding of peptidyl-tRNA stem to the donor site of the peptidyl-transferase center, stimulating the formation of inert complexes between the peptidyl end of peptidyl-tRNA and ribosomes, and perturbing the ribosome-donor stem complex (25, 26). Sparsomycin may increase −1 ribosomal frameshift efficiencies by either of two mechanisms: (i) a higher affinity of the donor stem for the ribosome may in itself slow down the rate of the peptidyl transfer reaction, or (ii) a change in the steric alignment between donor and acceptor tRNA stems may result in decreased peptidyl-transfer rates. Either of these two scenarios would increase the amount of time that a ribosome is paused over the frameshift signal with both its A- and P-sites occupied by tRNA, increasing the likelihood of −1 ribosomal frameshifting.
In the presence of either anisomycin or sparsomycin, the alterations in frameshifting efficiencies were significant enough to promote loss of L-A and M1 (Fig. 3C). The ability of these drugs to cure cells of L-A is significant. Of the 31 MAintenance Of Killer (mak) and 5 mof yeast mutations that cannot maintain M1, only three fail to propagate L-A as well (reviewed in ref. 5). Our results indicate that peptidyl-transferase inhibitors alter the efficiency of −1 programmed ribosomal frameshifting, consequently changing the ratio of available Gag to Gag-Pol, and ultimately interfere with the viral particle assembly process.
On the whole, the alteration in programmed frameshifting efficiency observed as a consequence of the drugs in cells was recapitulated in an in vitro translation system. Certain differences, however, between the in vitro and in vivo systems were observed. The effective concentrations of these drugs used to affect programmed frameshifting were three orders of magnitude lower in vitro than in whole cells, which is likely to be a reflection of drug uptake and metabolism in intact cells. Also, in the rabbit reticulocyte extract in vitro system, subnanomolar concentrations of the drugs stimulated luciferase production of the zero-frame control luciferase reporter, although the effects of the drugs on programmed −1 ribosomal frameshifting efficiencies followed the same general trends as observed in vivo. Finally, the baseline efficiency of −1 ribosomal frameshifting is significantly higher in vitro than in vivo. This could be indicative of differences in the stabilities of the reporter mRNAs in the two systems. Because the −1 ribosomal frameshift reporter mRNA has two in-frame termination codons within the first 200 bases of its 3.3-kb mRNA, it constitutes a nonsense-containing mRNA and has been demonstrated to be rapidly degraded in cells (15). The observed differences in the baseline efficiencies of −1 ribosomal frameshifting may be an indication that the nonsense-mediated mRNA decay pathway may be partially or wholly inactivated in the in vitro translation system (data not shown). Taken together, these factors are most likely responsible for higher baseline efficiencies of the −1 ribosomal frameshifting efficiency presented here.
The results presented here serve to further our understanding of programmed −1 ribosomal frameshifting by describing a refined conceptual framework, assays, and identifying compounds that affect this process. The ultimate goal of this research will be to identify antiviral agents. Future experiments will entail elucidating how these compounds alter these processes, and perhaps will allow us to develop better agents to fight against viral infections.
Acknowledgments
We thank Sidney Pestka for supplying anisomycin and sparsomycin and for helping us research antibiotics. We thank Terri Goss Kinzy, Thomas Thisted, Shuang Zhang, Carlos Gonzalez, and Amy Hammell for useful discussions and critical reading of the manuscript. This work was supported in part by grants given to J.D.D. by the Foundation of the University of Medicine and Dentistry of New Jersey and the State of New Jersey Commission on Cancer Research (96–62-CCR-00), and by grants awarded to S.W.P. by the National Institutes of Health (GM48631–01) and an American Heart Association Established Investigator Award.
ABBREVIATIONS
- dsRNA
double-stranded RNA
- mof
maintenance of frame
- aa-tRNA
aminoacyl-tRNA
- β-gal
β-galactosidase
References
- 1.Dinman J D. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farabaugh P J. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gesteland R F, Atkins J F. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 4.Dinman J D, Icho T, Wickner R B. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickner R B. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinman J D, Wickner R B. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinman J D, Wickner R B. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinman J D, Wickner R B. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson A J, Peltz S W. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 10.Leeds P, Peltz S W, Jacobson A J, Culbertson M R. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 11.Cui Y, Hagan K W, Zhang S, Peltz S W. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 12.He F, Jacobson A J. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 13.Weng Y, Czaplinski K, Peltz S W. Mol Cell Biol. 1996;16:5477–5490. doi: 10.1128/mcb.16.10.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng Y, Czaplinski K, Peltz S W. Genes Dev. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Y, Dinman J D, Peltz S W. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brierley I. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 17.Balasundaram D, Dinman J D, Wickner R B, Tabor C W, Tabor H. Proc Natl Acad Sci USA. 1994;91:172–176. doi: 10.1073/pnas.91.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallie D R, Feder J N, Schimke R T, Walbot V. Mol Gen Genet. 1991;288:258–265. doi: 10.1007/BF00282474. [DOI] [PubMed] [Google Scholar]
- 19.Fried H M, Fink G R. Proc Natl Acad Sci USA. 1978;75:4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leibowitz M J, Barbone F P, Georgopoulos D E. Methods Enzymol. 1991;194:536–545. doi: 10.1016/0076-6879(91)94040-j. [DOI] [PubMed] [Google Scholar]
- 21.Iizuka N, Najita L, Franzusorr A, Sarnow P. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belcourt M F, Farabaugh P J. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestka S. Molecular Mechanisms of Protein Biosynthesis. New York: Academic; 1977. pp. 467–553. [Google Scholar]
- 24.Carrasco L, Barbacid M, Vazquez D. Biochim Biophys Acta. 1973;312:368–376. doi: 10.1016/0005-2787(73)90381-x. [DOI] [PubMed] [Google Scholar]
- 25.Herner A E, Goldberg I H, Cohen L B. Biochemistry. 1969;8:1335–1344. doi: 10.1021/bi00832a006. [DOI] [PubMed] [Google Scholar]
- 26.Oen H, Pellegrini M, Cantor C R. FEBS Lett. 1974;45:218–222. doi: 10.1016/0014-5793(74)80848-3. [DOI] [PubMed] [Google Scholar]