Abstract
The yeast non-Mendelian genetic factor [PSI], which enhances the efficiency of tRNA-mediated nonsense suppression in Saccharomyces cerevisiae, is thought to be an abnormal cellular isoform of the Sup35 protein. Genetic studies have established that the N-terminal part of the Sup35 protein is sufficient for the genesis as well as the maintenance of [PSI]. Here we demonstrate that the N-terminal polypeptide fragment consisting of residues 2–114 of Sup35p, Sup35pN, spontaneously aggregates to form thin filaments in vitro. The filaments show a β-sheet-type circular dichroism spectrum, exhibit increased protease resistance, and show amyloid-like optical properties. It is further shown that filament growth in freshly prepared Sup35pN solutions can be induced by seeding with a dilute suspension of preformed filaments. These results suggest that the abnormal cellular isoform of Sup35p is an amyloid-like aggregate and further indicate that seeding might be responsible for the maintenance of the [PSI] element in vivo.
Self-propagating protein conformational changes have been proposed to be the cause of transmission of mammalian transmissible spongiform encephalopathies (1, 2), as well as for two yeast non-Mendelian inheritance elements, [PSI] and [URE3] (3–7). In the case of [PSI], an altered conformation of the Sup35 protein (Sup35p), which is the yeast homolog of the eukaryotic translation termination factor eRF3, is thought to be the determinant (3, 8). Consistent with this proposal it was observed that the maintenance of [PSI] only requires the N-terminal 114-amino acid domain of Sup35p, and that overproduction of Sup35p or its N-terminal fragment in yeast induces the de novo appearance of [PSI] (9–11). Differential sedimentation and fluorescence microscopic studies further established a correlation between Sup35p coalescence and the appearance of [PSI], suggesting that ordered aggregation converts newly synthesized Sup35p into its like and is thus responsible for the propagation of the [PSI] element (12, 13). In this study, we investigate the properties of the Sup35p polypeptide fragment consisting of residues 2–114, Sup35pN. It is found that Sup35pN aggregates to form amyloid-like filaments in vitro. We then show that seeding with preexisting filaments can speed up the formation of filaments from freshly prepared Sup35pN solutions.
EXPERIMENTAL PROCEDURES
Protein Purification and Filament Preparation.
A DNA fragment encoding the N-terminal 114-residue segment of Sup35p, with an extra Met–Gly–Ser2–His6–Ser2–Gly2–Ser segment at the N terminus and a stop codon at the C terminus, was obtained by PCR from yeast genomic DNA, using appropriate oligonucleotides. The fragment was inserted into the expression vector pMW172 (14) and the protein was overexpressed in the BLR21(DE3)/pLysS Escherichia coli strain (Novagen). Cell lysate was prepared in 20 mM Tris⋅HCl buffer (pH 7.9) containing 0.5 M NaCl, 5 mM imidazole, and 6 M guanidine hydrochloride (GdmCl), and loaded onto a Ni2+-NTA affinity column (Qiagen, Chatsworth, CA) followed by reverse-phase high performance liquid chromatography (HPLC, C18 column) in 0.1% trifluoroacetic acid (TFA) using an acetonitrile gradient. The N-terminal Met–Gly–Ser2–His6–Ser2–Gly2–Ser–Met segment was removed by cyanogen bromide cleavage (15). Unreacted peptide was separated from Sup35pN by a NaCl gradient, using an SP-Sepharose column (Pharmacia) in 20 mM sodium acetate (pH 4.5) containing 6 M urea, which was removed immediately after separation using a C18 reverse-phase HPLC column in 0.1% TFA with an acetonitrile gradient. Final buffer exchanges were achieved by gel filtration chromatography on Sephadex G25 columns (Pharmacia). The primary structure of Sup35pN was confirmed by electrospray mass spectrometry, sequencing of the N-terminal 25 residues, and isoelectric focusing. Filaments of Sup35pN were prepared in 0.1% (vol/vol) TFA/40% (vol/vol) acetonitrile using reverse-phase HPLC fractions containing isocratically eluted Sup35pN. A typical preparation of a 100 μM solution of Sup35pN yielded filaments after 1 week of incubation at 4°C. Spontaneous filament formation was also observed in 50 mM sodium phosphate buffer (pH 2.0) with 40% acetonitrile. No influence on filament formation was seen when borosilicate, polystyrol, or polypropylene tubes were used.
Electron Microscopy (EM).
Samples (5 μl) of the filament suspension were adsorbed to glow discharged carbon-coated copper grids. These were washed twice with deionized water, negatively stained with 2% (wt/vol) uranyl acetate, and air-dried after removal of excess liquid. The specimens were examined in a Philips CM12 transmission electron microscope at 100 kV, and images were recorded with a Gatan 694 slow scan CCD camera.
Circular Dichroism (CD) Spectroscopy.
CD spectra were recorded on a Jasco J710 spectropolarimeter at 22°C. All samples were briefly sonicated before measurement.
Polarization Light Microscopy.
Filaments were sedimented at 20,000 × g, washed with buffer A (50 mM sodium phosphate, pH 7.0/150 mM NaCl), stained with 50 μM Congo red in buffer A at room temperature for 1 min, washed in succession with buffer A and deionized water, then placed on glass slides, dried at room temperature, and covered. The samples were studied using a Zeiss Photomicroscope III equipped with crossed polars.
Protease K Resistance Assay.
About 8 μg of Sup35pN filaments were suspended in 18 μl Tris/EDTA (TE) buffer (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0) containing 0.8 M GdmCl. Two microliters of protease K (Boehringer Mannheim no. 1413 783) solution in TE buffer at concentrations from 0.5 to 4.0 μg/ml were added and the mixture was incubated at 37°C for 80 min. The reaction was terminated by adding 5 mM phenylmethylsulfonyl fluoride. Twenty microliters of SDS gel loading buffer (100 mM Tris⋅HCl, pH 6.8/4% SDS/0.2% bromophenol blue/20% glycerol) were added and the samples were boiled for 3 min. The resulting solution was briefly centrifuged and then subjected to 15% tricine–SDS/PAGE, which has particularly high resolution for small polypeptides (16). Protein was detected by Coomassie brilliant blue staining.
Seeding Assay.
Freshly prepared Sup35pN solutions were subjected to addition of different preparations of protein aggregates. In the case of seeding in 0.1% TFA/40% acetonitrile, a 50 μM solution of Sup35pN containing 1% (wt/wt) of protein aggregates as seeds was incubated at 4°C. In the case of seeding in 50 mM sodium phosphate at pH 7.0, a 5 μM solution of Sup35pN containing 5% of aggregates as seeds was incubated at 22°C. After overnight undisturbed incubation, aggregates were sedimented at 20,000 × g for 20 min, dissolved in 10 μl SDS gel loading buffer containing 6 M urea, boiled for 3 min, separated on 15% SDS polyacrylamide gels, and detected by Coomassie brilliant blue staining. For CD measurements, 2% (wt/wt) filaments were added as seeds to a freshly prepared 5 μM Sup35pN solution in 50 mM sodium phosphate at pH 7.0, and the mixture was incubated overnight at 22°C.
RESULTS
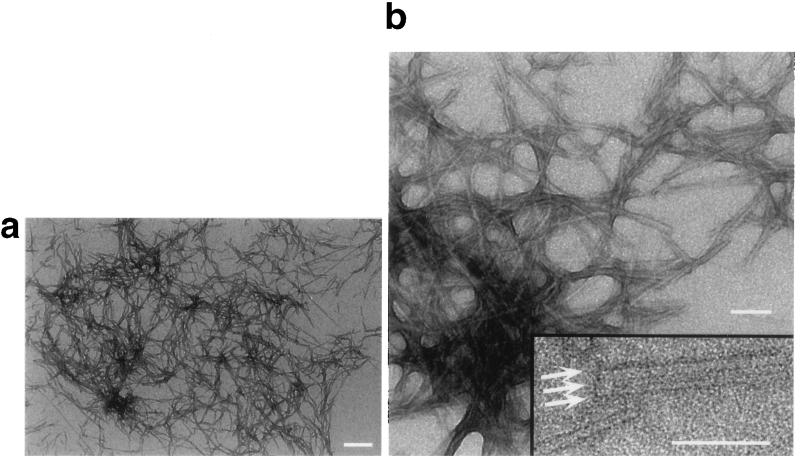
Sup35pN was overexpressed in E. coli and purified to homogeneity as judged by SDS/PAGE. It had limited solubility in aqueous buffers and, after sonication, submillimolar aqueous suspensions contained amorphous aggregates, as judged by EM. In 40% acetonitrile/60% H2O at pH 2.0 the formation of amorphous aggregates could be suppressed, and after prolonged incubation unbranched filaments with a diameter of 3 nm and variable lengths were observed (Fig. 1a). It was also noted that two or more filaments could further associate laterally to form fibrillar structures with larger diameter (Fig. 1b).
Figure 1.
(a and b) Electron micrographs of negatively stained Sup35pN filaments. (b Inset) Laterally associated fibrils are marked by arrows. (Bars = 250 nm in a and 100 nm in b and the Inset.)
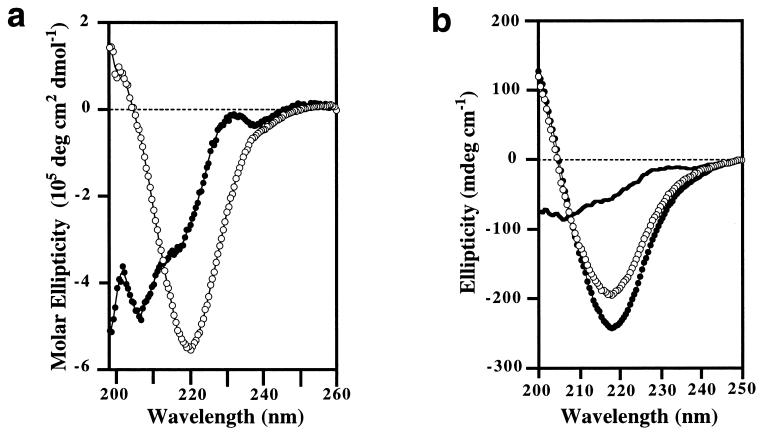
Sup35pN undergoes extensive secondary structural changes upon aging. A freshly prepared solution exhibits a far UV CD spectrum that indicates little α-helix or β-sheet content. In contrast, the spectrum of an aged solution shows β-sheet-like characteristics, with a single differential absorption minimum near 220 nm (Fig. 2a). These β-type spectral features are predominantly associated with the filaments, since the supernatant obtained after the filaments were removed from the aged solution by centrifugation had a similar CD spectrum as the freshly prepared Sup35pN solution (Fig. 2b). Amorphous aggregates of Sup35pN, which show strong light scattering, do not give CD signals. Freshly prepared and aged samples were indistinguishable by SDS/PAGE or isoelectric focusing under denaturing conditions (data not shown), indicating that the covalent polypeptide structure in the freshly prepared protein solution is preserved during aging.
Figure 2.
Far UV CD spectra of Sup35pN in 0.1% TFA/40% acetonitrile. (a) Normalized molar CD spectra of a freshly prepared sample (•) and an aged sample (○). (b) Ellipticity in a 40 μM aged Sup35pN solution (•) and its sedimented (○; resuspended for the CD measurement in 0.1% TFA/40% acetonitrile) and nonsedimented (solid line) parts after centrifugation at 20,000 × g. Small amounts of Sup35pN may have dissociated from the filaments during sedimentation and may contribute to the residual CD signal in the supernatant.
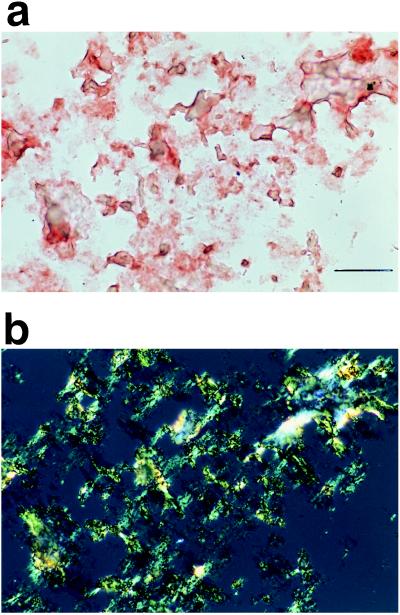
When Sup35pN filaments were collected by centrifugation and washed with sodium phosphate buffer at neutral pH, numerous red-stained aggregates were observed by light microscopy after Congo red treatment (Fig. 3a). Similar to amyloids, the stained aggregates exhibited green–yellow color when examined with crossed polars (Fig. 3b), showing that the filaments of Sup35pN are amyloid-like and contain regular secondary structure (17, 18).
Figure 3.
Photomicrographs of Sup35pN filaments stained with Congo red: (a) bright field, (b) polarized light. (Bar = 50 μm.)
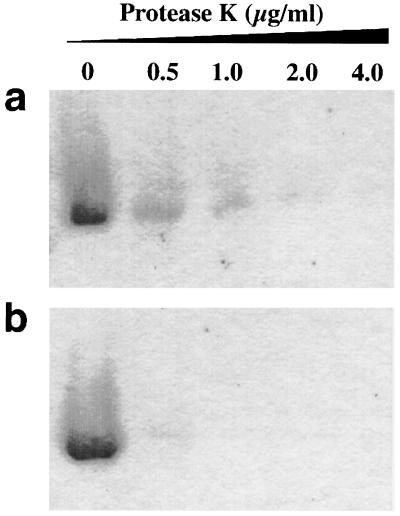
To test whether the in vitro generated filaments had increased protease resistance, we subjected the following two samples to protease K digestion. (i) Filaments were sedimented from an aged Sup35pN solution and resuspended in Tris/EDTA buffer (pH 8.0) containing 0.8 M GdmCl. (ii) As a control, the same number of filaments were denatured in 8 M GdmCl and then diluted 10-fold using Tris/EDTA buffer, whereby amorphous aggregation of denatured protein was induced by carrying out the dilution very slowly. The experiments showed that Sup35pN filaments have increased protease K resistance when compared with the control (ii) (Fig. 4).
Figure 4.
Protease K resistance of Sup35pN filamentous aggregates (a) and of amorphous aggregates (b) as determined by SDS/PAGE. The protease K concentration in each reaction is indicated.
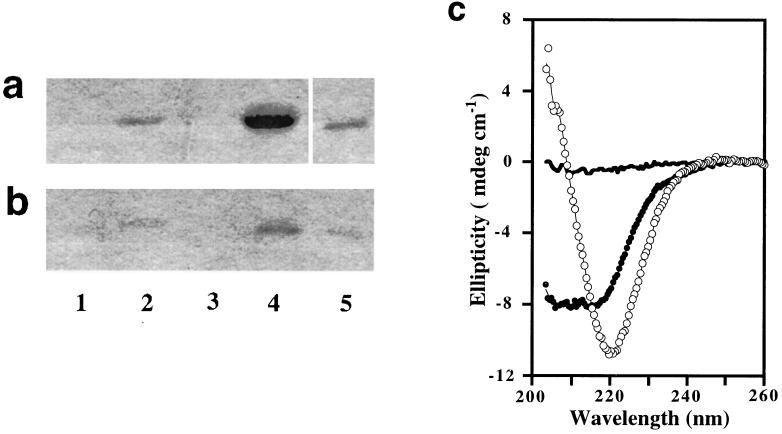
A key experiment in this investigation showed convincingly that seeding with filaments of Sup35pN leads to recruitment of more Sup35pN polypeptide into filaments. After addition of 1% (wt/wt) of the filaments to a freshly prepared solution of Sup35pN in 40% acetonitrile/60% H2O/0.1% TFA at pH 2.0 and overnight incubation at 4°C, large amounts of filamentous species could be isolated by centrifugation (Fig. 5a, lane 4) and identified by EM, while a mock-seeded solution yielded no sediments (Fig. 5a, lane 1). Formation of new filamentous structures was clearly associated with the addition of filaments, since the seeding activity was lost after denaturation of the filaments by 8 M GdmCl before they were used for seeding (Fig. 5a, lane 3). In another control, no increase of the degree of aggregation was seen within 1 day after seeding with the same amount of amorphous Sup35pN aggregates (Fig. 5a, lane 2).
Figure 5.
Gels showing filament growth after seeding in 0.1% TFA/40% acetonitrile (a) or in 50 mM sodium phosphate at pH 7.0 (b). Lane 1, mock-seeded; lane 2, seeding with amorphous aggregates; lane 3, seeding with GdmCl-denatured filaments; lane 4, seeding with filaments; lane 5, seeds alone directly loaded. (c) CD spectra of 5 μM Sup35pN in 50 mM sodium phosphate at pH 7.0: (•) mock-seeded, (○) seeded with filaments; the solid line corresponds to the spectrum of the added seeds.
Seeding was also observed at pH 7.0 in sodium phosphate buffer without organic cosolvent (Fig. 5b, lane 4). The association between seeding activity and presence of the filamentous structure was confirmed using the same controls as described above (Fig. 5b, lanes 1–4). This relation was further confirmed by comparison of the CD spectra of seeded and mock-seeded solutions after overnight incubation at 22°C and brief sonication. Fig. 5c shows that under these near-physiological solution conditions, seeding did also result in an increased β-type CD signal when compared with the mock-seeded reaction.
DISCUSSION
Previous work by others has established that Sup35 protein extracted from [PSI+] cells forms aggregates that exhibit partial protease resistance and interact with a recombinant Sup35p peptide fragment in vitro (12, 13). The presently described ordered filaments, which were prepared from a physiologically relevant peptide (9, 11), retain all the biochemical properties reported for ex vivo Sup35p aggregates. Although acidic pH and an organic cosolvent were initially used to suppress amorphous aggregation, once filaments appeared they were stable and capable of further growth also in aqueous solution at neutral pH.
The observed delayed filament formation in the absence of seeding and the seeding behavior of Sup35pN would be compatible with growth kinetics similar to those reported previously for amyloid fibrils, which elongate from critical nucleation sites of which the formation is rate-limiting (19–24). Such kinetics would offer an explanation for major features of [PSI] inheritance: spontaneous [psi−] to [PSI+] reversion is rare because de novo formation of an active nucleation site is difficult (10, 25); once such sites exist, propagation of the [PSI] determinant may involve recruiting newly synthesized peptide into filaments, which can release new nucleation sites (12). A similar mechanism was also proposed to rationalize the propagation of prions in mammalian species (refs. 19, 26, and 27; see also refs. 28 and 29 for alternative interpretations), where the infectious form of the prion protein is also an oligomer with amyloid characteristics (30). In this context, it is of further interest that EM studies of aged solutions of the prion-inducing domain of Ure2p (31), which is the cytosolic determinant of [URE3] (3, 7), also revealed filamentous structures (C.-Y.K. and K.W., unpublished work). The fact that mammalian prion proteins Sup35pN and Ure2p, which are all thought to undergo self-propagating conformation changes, are also able to adopt filamentous structures suggests that filament growth by seeding might be a common mechanism for prion propagation.
Acknowledgments
We thank Dr. A. Jancso for the expression plasmid; Dr. S. te Heesen for the yeast genomic DNA; Dr. G. Frank for peptide sequencing; Drs. S. te Heesen, R. Brunisholz, and Y. Chernoff for technical advice; Drs. R. Glockshuber and C. Weissmann for reading of the manuscript and helpful discussions; and Mrs. E. Ulrich for the careful processing of the manuscript. Financial support was obtained from the Human Frontier Science program, the Schweizerischer Nationalfonds (project 31.32033.91), and the Roche Foundation.
ABBREVIATIONS
- Sup35p
Sup 35 protein
- Sup35pN
Sup35 protein fragment of residues 2–114
- CD
circular dichroism
- TFA
trifluoroacetic acid
- GdmCl
guanidinium hydrochloride
- EM
electron microscopy
References
- 1.Prusiner S B. Trends Biol Sci. 1996;21:482–487. doi: 10.1016/s0968-0004(96)10063-3. [DOI] [PubMed] [Google Scholar]
- 2.Weissmann C. FEBS Lett. 1996;389:3–11. doi: 10.1016/0014-5793(96)00610-2. [DOI] [PubMed] [Google Scholar]
- 3.Wickner R B. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 4.Wickner R B, Masison D C, Edskes H K. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 5.Tuite M F. Nature (London) 1994;370:327–328. doi: 10.1038/370327a0. [DOI] [PubMed] [Google Scholar]
- 6.Cox B S, Tuite M F, McLaughlin C S. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 7.Lacroute F. J Bacteriol. 1971;106:519–522. doi: 10.1128/jb.106.2.519-522.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ter-Avanesyan M D, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 11.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 13.Patino M M, Liu J J, Glover J R, Lindquist S. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 14.Way M, Pope B, Gooch J, Hawkins M, Weeds A G. EMBO J. 1990;9:4103–4109. doi: 10.1002/j.1460-2075.1990.tb07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross E. Methods Enzymol. 1967;11:238–255. [Google Scholar]
- 16.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 17.Sipe J D. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 18.Glenner G G, Page D L. Int Rev Exp Pathol. 1976;15:1–92. [PubMed] [Google Scholar]
- 19.Jarrett J T, Lansbury P T., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 20.Lomakin A, Chung D S, Benedek G B, Kirschner D A, Teplow D B. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jarrett J T, Lansbury P T., Jr Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- 22.Come J E, Fraser P E, Lansbury P T., Jr Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colon W, Kelly J W. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 24.Wood S J, Maleeff B, Hart T, Wetzel R. J Mol Biol. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 25.Lund P M, Cox B S. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 26.Brown P, Goldfarb L G, Gajdusek D C. Lancet. 1991;337:1019–1022. doi: 10.1016/0140-6736(91)92670-w. [DOI] [PubMed] [Google Scholar]
- 27.Caughey B, Kocisko D A, Raymond G J, Lansbury P T., Jr Curr Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 28.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 29.Wille H, Zhang G-F, Baldwin M A, Cohen F E, Prusiner S B. J Mol Biol. 1996;259:608–621. doi: 10.1006/jmbi.1996.0343. [DOI] [PubMed] [Google Scholar]
- 30.DeArmond S J, McKiniey M P, Barry R A, Braunfeld M B, McColloch J R, Prusiner S B. Cell. 1985;41:221–235. doi: 10.1016/0092-8674(85)90076-5. [DOI] [PubMed] [Google Scholar]
- 31.Masison D C, Wickner R B. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]