Abstract
The proteasome is a multicatalytic protease complex that plays a key role in diverse cellular functions. The peptide vinyl sulfone, carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone (Z-L3VS) covalently inhibits the trypsin-like, chymotrypsin-like and, unlike lactacystin, also the peptidylglutamyl peptidase activity in isolated proteasomes, and blocks their function in living cells. Although described as a class of mechanism-based inhibitors for cysteine proteases, the peptide vinyl sulfone Z-L3VS and a 125I-labeled nitrophenol derivative (125I-NIP-L3VS) covalently modify the active site threonine of the catalytic β subunits of the proteasome. Modification of Thermoplasma proteasomes demonstrates the requirement for a hydroxyl amino acid (threonine, serine) as nucleophile at the β subunit’s NH2 terminus. 125I-NIP-L3VS covalently modifies the HslV subunit of the Escherichia coli protease complex HslV/HslU, a reaction that requires ATP, and supports a catalytic mechanism shared with that of the eukaryotic proteasome.
Proteasomes are multicatalytic proteolytic complexes found in almost all living cells and are responsible for the degradation of the majority of cytosolic proteins in mammalian cells (1). The 20S proteasome is a 700-kDa barrel-shaped structure of four stacked rings (2) that contains two types of subunits; α subunits make up the outer two rings of the complex and the catalytic β subunits the inner two rings. Proteasomes of the archaebacterium, Thermoplasma acidophilum are comprised of 14 identical α and 14 identical β subunits. Eukaryotic proteasomes contain seven different but homologous α subunits, and the β rings contain seven distinct but related β subunits (20S proteasome) (3). The 20S proteasome is the catalytic core of the larger, ATP-dependent, 26S complex that is responsible for the degradation of ubiquitin-conjugated proteins (4). Further complexity arises from the possible replacement of the catalytic β subunits X, Y, and Z with the interferon-γ-inducible, major histocompatibility complex (MHC)-encoded subunits LMP-2, LMP-7, and with MECL-1 (5).
Initial attempts to classify the proteasome’s catalytic mechanism into a category with known proteases were unsuccessful mainly due to a lack of homology with known peptidases (6). Mutational and structural studies uncovered a novel catalytic mechanism, involving an NH2-terminal threonine residue as the catalytic nucleophile (2, 7): the free amino terminus or the ɛ amino group from a conserved, nearby lysine residue activates the threonine hydroxyl group for nucleophilic attack on the peptide bond (7).
The ubiquitin proteasome pathway is involved in many diverse cellular functions including cell cycle progression, antigen presentation, and activation of transcription factors (8–10). Inhibitors of the proteasome are thus of interest as tools for studying proteasomal involvement. Peptide aldehydes are potent, reversible inhibitors that inactivate the proteasome’s multiple active sites by forming a transient, covalent hemiacetal with the catalytic NH2-terminal threonine hydroxyl (9, 11). Peptide aldehydes are active against proteasomal proteolysis both in vitro and in intact cells but can also inhibit cellular thiol proteases that can complicate the interpretation of certain studies (1, 11).
Lactacystin is an irreversible, covalent inhibitor of the chymotrypsin-like and trypsin-like activities and a weak, reversible inhibitor of the peptidylglutamyl peptidase activity of the proteasome (12). Its exquisite specificity has made lactacystin a useful reagent for studying proteasome function in mammalian cells, but its modest activity against proteasomes from archaebacteria and against certain eubacterial homologs has limited its use in studies of these related enzymes.
We report here a new class of inhibitors of the proteasome: peptide vinyl sulfones. The vinyl sulfone acts as a Michael acceptor for soft nucleophiles such as thiols, leading to the formation of a covalent bond (13) (Fig. 1B), while resistant to attack by free thiols that can cause inactivation of other classes of protease inhibitors (13). We show that the tripeptide vinyl sulfone Z-L3VS and related derivatives quite unexpectedly inhibit the trypsin-like, the chymotrypsin-like, and, unlike lactacystin, the peptidylglutamyl peptidase activity of the proteasome in vitro by covalent modification of the NH2-terminal threonine of the catalytically active β subunits. They are more easily synthesized than lactacystin and can be conveniently tagged with either biotin for purposes of affinity chromatography (M.B. and H.P., unpublished observation), or a nitrophenol moiety for subsequent radiolabeling. We show that a 125I-labeled vinyl sulfone of the tripeptide sequence Leu-Leu-Leu selectively modifies β subunits in purified proteasome preparations as well as in whole cell homogenates and in living cells of widely different origin.
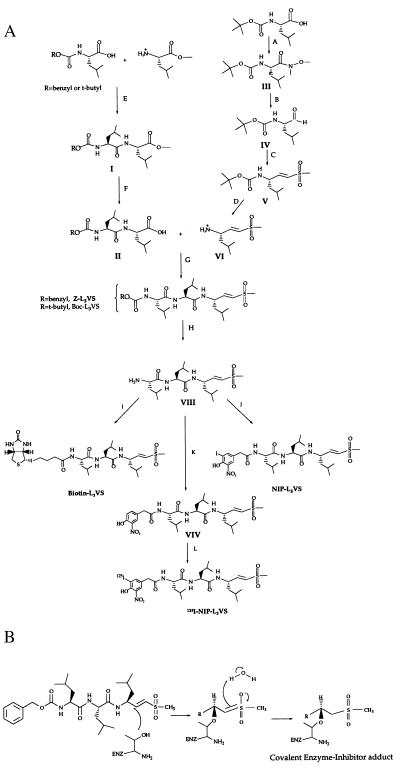
Figure 1.
Synthesis (A) and mechanism of inhibition (B) of the peptide vinyl sulfones. (A) PyBOP, DIEA, CH3NHOCH3, CH2Cl2; (B) LAH, Et2O (C) (EtO)2P(O)CH2S(O)2CH3, NaH; TsOH, Et2O; (E) DCC, HOBT, DMF; (F) 20% K2CO3/MeOH; (G) PyBOP, DIEA, CH2Cl2; (H) TsOH, Et2O (I) Biotin-NP ester, DMF, DIEA; (J). 4-Hydroxy-3-iodo-5-nitrophenylacetic acid, PyBOP, DIEA, CH2Cl2; (K) 4-hydroxy-3-nitrophenylacetic acid, PyBOP, DIEA, CH2Cl2; (L) Na125I, Iodogen.
Recently an ATP-dependent protease complex comprised of two heat shock proteins has been discovered (14). The HslV gene product is a peptidase that shows homology to β type subunits of the proteasome, including the presence of a NH2-terminal threonine (15, 14). HslV is cotranscribed with the adjacent HslU gene that encodes an ATPase with homology to other known Escherichia coli ATPases such as ClpX (50% identity) (16). Together the HslV and HslU gene products make up a complex with an ATP-dependent proteolytic activity similar to that of the eukaryotic proteasome (14). We show that peptide vinyl sulfones covalently modify HslV only in the presence of HslU and ATP, consistent with the reported nucleotide dependence of the activity of this complex (14). These observations provide experimental support for the HslU/HslV complex’s proposed functional homology to the proteasome and indicate that ATP influences the formation of the active site of this enzyme complex.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture.
The human cell lines HOM-2, T2, and US11 transfectants prepared from the astrocytoma cell line, U373-MG have been described (20). The US11 transfectants were cultured in DMEM supplemented with 10% fetal calf serum and puromycin (Sigma) at a final concentration of 0.375 μg/ml. All other cells were grown in RPMI media 1640 supplemented with 10% fetal calf serum.
Antibodies.
Proteasomes were immunoprecipitated using a mAb that recognizes an α subunit as part of the mature 720-kDa proteasome complex (Organon Teknika, Turnhout, Belgium) (24). MHC class I molecules were immunoprecipitated using a rabbit anti-class I heavy chain serum (25).
Biochemical Methods and Materials.
Peptide inhibitors (Z-L3H, Z-L3VS, and NIP-L3VS) were dissolved in dimethyl sulfoxide and diluted in cell culture media to the desired concentration, keeping the final concentration of dimethyl sulfoxide at <1%. Cells (1.5 × 106), purified proteasomes (0.1 μg for rabbit, Saccharomyces cerevisiae, and T. acidophilum; 0.1 μg HslV and 0.4 μg HslU) or lysates (100 μg of total protein) were labeled with 125I-NIP-L3VS, diluted to a final concentration of 1.8 × 104 Bq/ml in tissue culture medium (cells) or reaction buffer (50 mM Tris, pH 7.4/2 mM dithiothreitol/5 mM MgCl2/2 mM ATP/purified proteasomes), at 37°C for 2 hr. For labeling in the absence of ATP, apyrase (5 unit/ml) was used instead of ATP. Labeling of cells was quenched by washing three times with PBS followed by addition of 1× SDS-Laemmli sample buffer. Labeling of purified proteasomes and total lysates was quenched by addition of 4× Laemmli sample buffer (to 1×). All samples were resolved by SDS/PAGE, nonequilibrium pH gradient gel electrophoresis/PAGE, or isoelectric focusing/PAGE. Preparation of lysates and immunoprecipitations was performed as described (25).
Kinetics of Inhibition of Proteasomal Peptidase Activity.
Purified mixtures of 20S and 26S proteasomes were isolated from U373-MG cells by differential centrifugation and anion-exchange chromatography as described (5, 26). 20S proteasomes were separated from these mixtures by native PAGE (4% polyacrylamide gel). Mixtures of 20S and 26S proteasomes or pure complexes were used to determine kinetics of inhibition of the proteasomal peptidases. Fluorogenic peptide substrates (100 μM final concentration) were used as substrates for the hydrophobic (Suc-LLVY-MCA), basic (Boc LRR-MCA), and acidic (Cbz-LLE-βNA) peptidase activities as described (5, 26). The rates of association (Kassoc) of Z-L3VS were determined as described (12).
Synthesis of the Peptide Vinyl Sulfones.
Peptide vinyl sulfones were synthesized on a 5–10 mmol scale following the strategy outlined in Fig. 1A, which, in essence, corresponds to the scheme described by Palmer (13). Details of the synthetic procedures are available from the authors upon request.
Pulse–Chase Analysis.
Cells were detached by trypsin treatment and incubated in methionine-free DMEM with or without added inhibitors for 1 hr. Two million cells (10 min pulse; 250 μCi of label; 1 Ci = 37 GBq) were used per sample. Incorporation was terminated by the addition of nonradioactive methionine to a final concentration of 1 mM. Immediately after the chase, samples were placed on ice and lysed in 1 ml of ice-cold Nonidet P-40 lysis mix. Preparations of lysates and immunoprecipitations were performed as described (25).
Gel Electrophoresis.
SDS/PAGE, two-dimensional isoelectric focusing/PAGE, fluorography, and autoradiography were performed as described (27, 20). Two-dimensional nonequilibrium pH gradient gel electrophoresis/PAGE was performed as described (28).
RESULTS
Synthesis of Peptide Vinyl Sulfones.
Tripeptide vinyl sulfones were synthesized containing the sequence Leu-Leu-Leu (Z-L3VS, Fig. 1A). Analogs of Z-L3VS in which the carboxybenzyl group was replaced with biotin (Biotin-L3VS), the hapten 4-hydroxyl-5-iodo-3-nitrophenyl acetate (NIP-L3VS), and the corresponding radiolabeled hapten (125I-NIP-L3VS) were also synthesized.
Peptide Vinyl Sulfones Irreversibly Inhibit Proteasome Activity in Vitro.
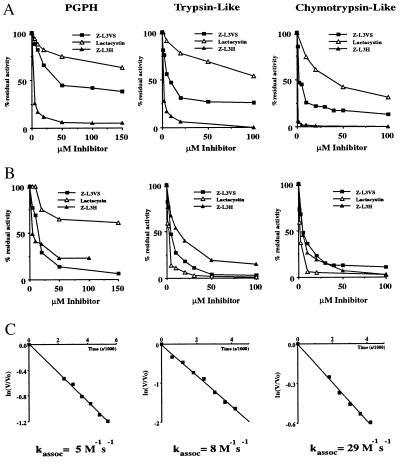
The ability of the peptide vinyl sulfones to inhibit proteasomal proteolysis was determined in vitro using a partially purified mixture of 20S and 26S proteasomes with fluorogenic peptides as substrates. Strongest inhibition was observed for the chymotrypsin-like peptidase activity of the proteasome with somewhat lesser inhibition of the trypsin-like and peptidylglutamyl peptidase activities, as shown for Z-L3VS (see Fig. 2). The peptide vinyl sulfones, including the biotin and NIP derivatives (data not shown), were only slightly less potent inhibitors than the peptide aldehyde Z-L3H (Fig. 2A) and were more active toward all three peptidase activities than was lactacystin upon simultaneous addition with substrates. When each of the three classes of inhibitors was preincubated for 1 hr with proteasomes and then diluted 10-fold, the extent of inhibition by the aldehyde was reduced, as expected for a reversible inhibitor, while the extent of inhibition by both lactacystin and Z-L3VS increased, consistent with time-dependent, irreversible inhibition (Fig. 2B). Furthermore, Z-L3VS caused inhibition of the trypsin-like peptidase activity to a greater extent than lactacystin (Fig. 2B), indicating that Z-L3VS binds to a subunit or binding site more effectively than lactacystin.
Figure 2.
The peptide vinyl sulfones inhibit the proteasome in vitro. Purified preparations of 20S and 26S proteasomes were incubated with increasing concentrations of the three inhibitors [Z-L3H, Z-L3VS, and lactacystin either simultaneously with substrates (A)], or samples were preincubated for 1 hr (37°C) and diluted 10-fold prior to addition of substrates (B). Hydrolysis of substrates corresponding to the acidic (Cbz-LLE-βNA), basic (Boc-LRR-MCA), and hydrophobic (Suc-LLVY-MCA) activities of the proteasome were measured by fluorescence spectroscopy. Kassoc for the compound, Z-L3VS, were determined for the three peptidase activities of purified 20S proteasomes (C). V represents velocity of the reaction at the time (t), and Vo is the velocity at time 0. Kassoc = ln(V/Vo)/[I] where [I] is the concentration of inhibitor.
125I-NIP-L3VS Covalently Modifies Multiple β Subunits in Purified Proteasomes Preparations and in Total Cell Extracts.
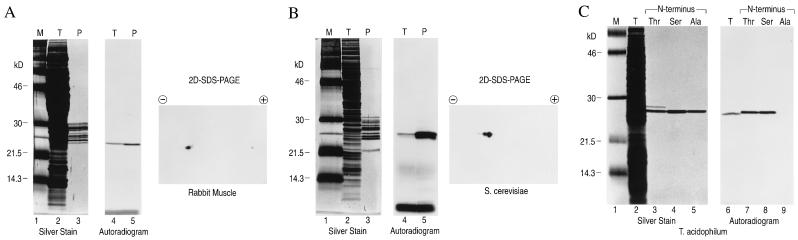
A radiolabeled version of the peptide vinyl sulfone NIP-L3VS was used to examine the ability of this class of compounds to react with proteasomes of diverse origin. 125I-labeled NIP-L3VS was incubated with purified 20S proteasomes and with total cell homogenates from rabbit muscle and yeast (Fig. 3 A and B). A single band of 23 kDa was labeled both in crude cell extracts and in purified proteasomes. However, in both cases this band could be resolved into at least two polypeptides of similar molecular weight but different isoelectric points.
Figure 3.
The peptide vinyl sulfone, 125I-NIP-L3VS, labels multiple β subunits of the proteasome in total cell extracts and in purified proteasome preparations. Total cell extracts (T) and purified preparations of proteasomes (P) were incubated with 125I-NIP-L3-VS (1.8 × 104 Bq/ml) at 37°C for 2 hr, resolved on a 12.5% polyacrylamide gel and visualized by silver staining, by autoradiography or by two-dimensional isoelectric focusing SDS/PAGE for rabbit muscle (A), S. cerevisiae (B), or T. acidophilum (C). For T. acidophilum, recombinant β subunits containing an NH2-terminal threonine, serine, or alanine were compared, as indicated.
The Peptide Vinyl Sulfones Are Reactive Against the Archaebacterial Proteasome from Thermoplasma.
Recombinant mutant and wild-type proteasome β subunits from the archaebacterium, Thermoplasma, were used to directly examine whether the NH2-terminal threonine is required for covalent modification by a peptide vinyl sulfone. Labeling of total cell lysates from Thermoplasma was compared with three preparations of recombinant proteasomes in which the catalytic NH2-terminal threonine was replaced with either serine or alanine. Recent studies have shown that β subunits in which the NH2-terminal threonine is replaced by serine retain full activity against peptide substrates, while replacement with alanine leads to complete loss of peptidase activity (7). 125I-NIP-L3VS labeled not only the β subunits in total extracts but also in the mutant complex in which the NH2-terminal threonine is replaced by serine (Fig. 3C). The NH2-terminal alanine mutant was refractory to labeling, indicating that covalent modification of β subunits requires the catalytic NH2-terminal threonine of the proteasome. The slight difference in apparent molecular weight between the β subunits from total cell extracts and the recombinant forms of β subunits is due to the presence of a histidine tag.
The Bacterial Protease Complex HslV/HslU from E. coli Is a Target for the Peptide Vinyl Sulfones.
The proteasome-like protease complex from E. coli is composed of two types of heat shock protein subunits, HslV and HslU (14). The β-like subunit HslV contains an NH2-terminal threonine, but direct evidence for a catalytic role for this residue is lacking (15). To address the catalytic mechanism of this complex, purified preparations of HslV/HslU were treated with the 125I-labeled peptide vinyl sulfone. 125I-NIP-L3VS covalently modified HslV, but only in the presence of HslU and ATP (Fig. 4), consistent with the reported requirement for ATP of the HslV/HslU complex (14). Combined, our results show that the peptide vinyl sulfones are capable of inhibiting proteases of the proteasome family found in a wide variety of species, including the eubacterial species E. coli, by targeting the catalytic, active site threonine residue.
Figure 4.
The peptide vinyl sulfones covalently modify the HslV gene product from the E. coli protease complex HslV/HslU. Purified preparations of HslV and HslU were incubated with 125I-NIP-L3VS (1.8 × 104 Bq/ml) independently or as a 1:4 mixture (HslV:HslU) in the presence of 2 mM ATP (+ATP) or 5 unit/ml apyrase (−ATP).
125I-NIP-L3VS Covalently Modifies β Subunits of the Proteasome in Living Cells in a Time- and Dose-Dependent Manner.
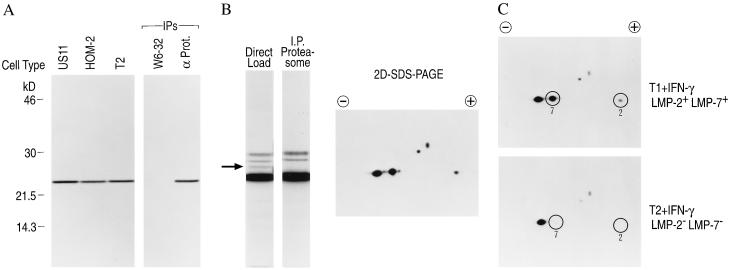
The ability of the tripeptide vinyl sulfones to penetrate cells and specifically modify proteasomal β subunits was established using several different cell lines (Fig. 5A). Incubation of 125I-NIP-L3VS with the human astrocytoma cell line U373-MG, the human B-cell line HOM-2, and the human T2 cell line (17) lacking the two interferon-inducible β subunits LMP-2 and LMP-7 showed labeling of a 23-kDa species similar to that described for rabbit muscle preparations. The labeling increased linearly with time and with concentration of 125I-NIP-L3VS (data not shown). The labeled polypeptides could be immunoprecipitated using a mAb raised against an α subunit common to the 20S and 26S proteasome, but not with an irrelevant antibody, thus establishing that the labeled polypeptides are part of the proteasome complex.
Figure 5.
The peptide vinyl sulfones covalently modify multiple β subunits of the proteasome in living cells including the γ-interferon-inducible subunits LMP-2 and LMP-7. (A) US11+, HOM-2, and T2 cells (1.5 × 106 cells) were incubated with 125I-NIP-L3VS (1.8 × 104 Bq/ml) for 2 hr at 37°C, washed several times, and lysed. Total cell extracts were resolved on a 12.5% polyacrylamide gel and visualized by autoradiography. Immunoprecipitations were performed on lysates from 125I-NIP-L3VS-labeled HOM-2 cells using mAbs recognizing MHC class I molecules (W6-32) or a proteasomal α subunit (α prot.) (B) HOM-2 cells were incubated with 125I-NIP-L3VS, lysed, and analyzed by autoradiography (Direct Load) or proteasomes were immunoprecipitated (I.P. Proteasome) as in A. Note the absence of a single labeled polypeptide (26 kDa) in the proteasome immunoprecipitation. A total cell lysate from 125I-NIP-L3VS labeled HOM-2 cells was also separated by two-dimensional nonequilibrium pH gradient gel electrophoresis/PAGE (11% polyacrylamide gel) and visualized by autoradiography. (C) T1 and T2 cells (1.5 × 106 cells) were incubated with 125I-NIP-L3VS (1.8 × 104 Bq/ml) for 2 hr at 37°C, lysed, resolved by two-dimensional nonequilibrium pH gradient gel electrophoresis/PAGE, and analyzed by autoradiography. Note the presence of labeled LMP-2 and LMP-7 in lysates from T1 cells only.
The identity of the proteasome β subunits modified by the radiolabeled vinyl sulfone was determined using the mutant human cell line, T2, in which the MHC region coding for the two interferon-inducible proteasome β subunits, LMP-2 and LMP-7, has been deleted (17). We could thus determine that two of the major species labeled with 125-I-NIP-L3VS were LMP-2 and LMP-7 by comparison with the wild-type cell line T1 (Fig. 5C). The most intensely labeled, basic polypeptide corresponding to the subunit X was present in similar quantities in both cell lines, as were the two minor species of higher molecular weight. The more acidic polypeptide was absent in the labeling of T2 cells. Based on the known pI values of the β subunits (1), the latter spot was identified as LMP-2. Similarly, a polypeptide of a molecular weight identical to X but with a slightly more acidic pI was present in the T1 cells and absent from T2 cells, identifying it as the LMP-7 subunit of the proteasome. These assignments were confirmed by comparison of mouse LMP-2 and LMP-7 obtained from different H2 haplotypes, where these subunits can be resolved based on differences in their molecular weight and isoelectric point (18) (data not shown).
Upon longer exposure of autoradiograms from the labeled cell extracts, three less intensely labeled polypeptides of 26, 28, and 29 kDa were visible (Fig. 5B). Two of these species were coimmunoprecipitated with the predominant 23-kDa band when using a proteasome-specific antibody (Fig. 5B). The third, minor species is a nonproteasomal protease modified to some extent by the peptide vinyl sulfones (see below). Analysis by two-dimensional gel electrophoresis (Fig. 5B) shows that the predominantly labeled, 23 kDa band is composed of three polypeptides of different isoelectric points, while the higher molecular weight species each represent individual subunits.
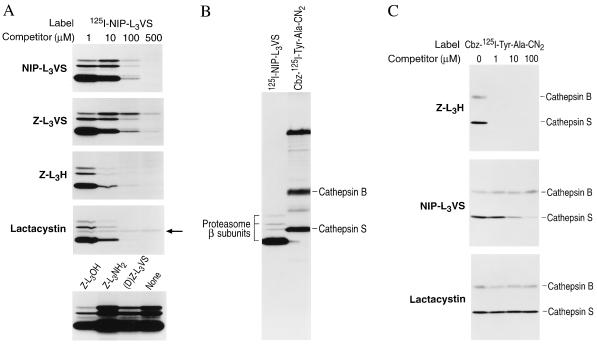
All Three Classes of Proteasome Inhibitors Compete for Binding with 125I-NIP-L3VS.
To address the specificity of the radiolabeled inhibitor for the proteasome, competition experiments were performed on intact mammalian cells (Fig. 6A). The peptide aldehyde Z-L3H blocks covalent binding of the labeled inhibitor to all β subunits at a concentration of 10 μM. The compounds NIP-L3VS, as well as the structurally related compound, Z-L3VS, also competitively reduced subunit modification by 125I-NIP-L3VS but were slightly less effective than the aldehyde Z-L3H and lactacystin. The tripeptide vinyl sulfone in which the COOH-terminal leucine is replaced with the D-isomer [(D)Z-L3VS] did not compete for binding even at 500 μM.
Figure 6.
The peptide vinyl sulfones, the peptide aldehyde, and lactacystin compete for binding to all proteasome subunits modified by 125I-NIP-L3VS. (A) HOM-2 cells were incubated simultaneously with 125I-NIP-L3VS (1.8 × 104 Bq/ml) and with increasing concentrations of NIP-L3VS, Z-L3VS, lactacystin, or with 500 μM of the control peptides Z-L3OH, Z-L3NH2, or (D)Z-L3VS for 2 hr at 37°C. Note the 26-kDa polypeptide shown in Fig. 5B remains resistant to competition by lactacystin (see ←). (B) HOM-2 cells were labeled with 125I-NIP-L3VS as described or with the cysteine protease inhibitor Cbz-125I-Tyr-Ala-CN2 for 2 hr at 37°C. Lysates were separated on a 12.5% polyacrylamide gel and analyzed by autoradiography. (C) HOM-2 cells were incubated simultaneously with Cbz-125I-Try-Ala-CN2 and increasing concentrations of Z-L3H, NIP-L3VS, and lactacystin as indicated for 2 hr at 37°C. Lysates were resolved on a 12.5% polyacrylamide gel and analyzed by autoradiography.
Of the species modified by the radiolabeled vinyl sulfone, only one rather minor polypeptide (seen only in mammalian cells) was resistant to competition by lactacystin (Fig. 6A, ←). Because peptide aldehydes such as Z-L3H are known to inhibit other classes of proteases (1, 11), while lactacystin does not, we presumed this minor polypeptide to be a nonproteasomal protease.
To identify the lactacystin-resistant, 26-kDa species, cells were labeled with the cysteine protease-selective probe, Cbz-125I-Tyr-Ala-N2 (Fig. 6B). This modified dipeptide covalently modifies cysteine proteases of the cathepsin family, predominantly cathepsins B and S (19). Comigration of the weakly labeled 26-kDa species with the polypeptide identified as the lysosomal protease cathepsin S confirmed its identity. We further examined the ability of all three classes of proteasome inhibitors to compete with the Cbz-125I-Tyr-Ala-CN2 probe for binding to cathepsins B and S (Fig. 6C). The peptide aldehyde, Z-L3H, was able to prevent labeling of both cathepsin S and B at concentrations as low as 1 μM, while lactacystin showed no inhibition of labeling of either, at concentrations as high as 100 μM. NIP-L3VS showed only partial inhibition of labeling of cathepsin S and no inhibition of labeling of cathepsin B even at the two highest concentrations tested. This result strongly suggests that Z-L3H could, in principle, block lysosomal proteases involved in antigen presentation.
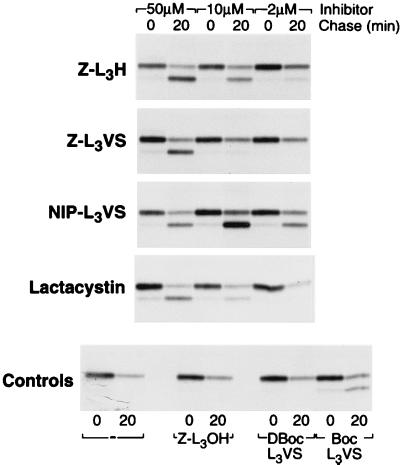
Peptide Vinyl Sulfones Inactivate the Proteasome In vivo.
The in vivo effects of the peptide vinyl sulfones were compared with the other two classes of proteasome inhibitors Z-L3H and lactacystin (Fig. 7). The peptide vinyl sulfones were less toxic to cells than the peptide aldehyde Z-L3H as assessed by their inhibitory affects on protein synthesis (data not shown). Proteasomal inhibition was examined using an assay in which a viral protein promotes destruction of MHC class I heavy chains in a proteasome-dependent manner (20) (21). The human cytomegalovirus gene US11 causes the dislocation of free class I heavy chains from the ER to the cytosol, at which point the single N-linked glycan is removed and the class I heavy chain is degraded by the proteasome. Thus, in the presence of US11 and an active proteasome inhibitor, the transient appearance of a cytosolic, deglycosylated breakdown intermediate results (20) (21). Z-L3VS caused the characteristic build-up of this heavy chain intermediate. The peptide vinyl sulfone analog containing the hapten NIP at the NH2 terminus showed activity at 2 μM, better than that of lactacystin, Z-L3H and the related compound, Z-L3VS. The peptide vinyl sulfone in which the COOH-terminal leucine was replaced with the D-isomer (DBocL3VS) was inactive in this assay.
Figure 7.
The peptide vinyl sulfones block proteasome function in vivo. US11+ cells were preincubated in methionine-free medium without (−) or with the indicated concentrations of Z-L3H, Z-L3VS, NIP-L3VS, or lactacystin for 1 hr. Cells were pulse labeled for 10 min and chased for 20 min. Class I molecules were immunoprecipitated with rabbit anti-heavy chain serum, resolved on a 12.5% polyacrylamide gel, and analyzed by fluorography. Note the presence of a class I heavy chain band devoid of carbohydrates, which migrates faster than the glycosylated species, and accumulates in the presence of an active proteasome inhibitor. ZL3OH, peptide alcohol.
DISCUSSION
Inhibitors of the proteasome have been instrumental in deciphering its role in a diverse assortment of cellular processes (9, 22, 10). We report here a class of inhibitors, the peptide vinyl sulfones, which inactivate the proteasome by covalent modification of the active site threonine of the catalytic β subunits of widely different origin. The ability of the peptide vinyl sulfones to permeate cells and covalently modify proteasome β subunits also makes them useful active site probes for studying the function of the proteasome in living cells.
Although originally characterized as mechanism-based inhibitors of cysteine proteases, the peptide vinyl sulfones inhibit the chymotrypsin-like, trypsin-like, and peptidylglutamyl peptidase activities of the proteasome both in vitro and in vivo. The activity of the vinyl sulfones against a side chain threonine hydroxyl nucleophile is surprising considering that these types of Michael acceptor structures were designed as mechanism-based inhibitors for soft nucleophiles such as cysteine (13). The peptide vinyl sulfones therefore provide new information about the reactivity of a threonine residue as an active site nucleophile. The ability of the vinyl sulfone to covalently modify a side chain hydroxyl of serine when placed at the NH2 terminus of a proteasomal β subunit, but not when part of the active site of a serine protease, suggests basic differences in the way the proteasome and serine proteases utilize a hydroxyl group as a catalytic nucleophile.
The reactivity of the peptide vinyl sulfones against the ATP-dependent eubacterial proteasome homolog, HslU/HslV, provides solid evidence that this multicomponent protease complex uses a catalytic mechanism similar to that of the proteasome. Moreover, modification required the presence of the peptidase subunit (HslV), the regulatory ATPase (HslU), and ATP, all of which are necessary for the cleavage of a peptide substrate. These findings suggest that the vinyl sulfones inactivate their target via a mechanism resembling that involved in peptide hydrolysis.
Competition experiments using the radiolabeled peptide vinyl sulfone was used to assess the activity and specificity of three different classes of proteasome inhibitors. The inability of the tripeptide vinyl sulfone containing a d-leucine in the P1 position to inhibit labeling of any of the β subunits labeled by 125I-NIP-L3VS indicated the proteasome’s stereochemical selectivity at the site of hydrolysis. The labeling of a rather minor 26-kDa polypeptide, identified as cathepsin S, was completely blocked by Z-L3H but was unaffected by lactacystin at all concentrations tested. This finding, combined with the competition data obtained for Z-L3H, NIP-L3VS, and lactacystin using the 125I-Tyr-Ala probe, fits not only the known specificity of lactacystin for the proteasome but also suggests that the peptide aldehyde, Z-L3H, should be a potent competitive inhibitor of lysosomal proteases. Related peptide aldehydes are known to inhibit lysosomal and Ca2+-dependent cysteine proteases, and such possible effects must be considered in studies using these agents on intact cells (1, 11).
The finding that lactacystin competes for labeling of all five β subunits by 125I-NIP-L3VS, is inconsistent with the initial identification of a single β subunit, X, as the target of the drug (12). While the predominant species modified by lactacystin is X, modification of other subunits that require higher concentrations of drug may have escaped detection due to the relatively low concentration of labeled compound used, as confirmed in subsequent work (23). We conclude that 125I-NIP-L3VS labels at least five distinct β subunits, all of which are also targets for lactacystin. Thus, the peptide vinyl sulfones can be used to provide detailed information on proteasomal inhibition by other classes of compounds. Their ease of synthesis and ability to covalently inactivate proteasomal β subunits in intact cells makes peptide vinyl sulfones useful tools for the study of proteasome function.
Acknowledgments
We thank Markus Rohrwild and H.-C. Huang for purified HslV/HslU, Peter Zwickl for Thermoplasma lysates, Alexei Kisselev for purified preparations of recombinant Thermoplasma proteasomes, and Kee Min Woo for purified 20S proteasomes from rabbit muscle. We also thank Evan Powers (Massachusetts Institute of Technology Chemistry Department) for helpful suggestions and technical assistance in the synthesis of these inhibitors. This work was supported by funding from the National Institutes of Health.
ABBREVIATIONS
- Z-L3VS
carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone
- 125I-NIP-L3VS
125I-labeled nitrophenol derivative
- MHC
major histocompatibility complex
References
- 1.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 2.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 3.Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 5.Gaczynska M, Rock K L, Goldberg A L. Nature (London) 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 6.Zwickl P, Grziwa A, Pühler G, Dahlmann B, Lottspeich F, Baumeister W. Biochemistry. 1992;31:964–972. doi: 10.1021/bi00119a004. [DOI] [PubMed] [Google Scholar]
- 7.Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 8.Ghislain M, Udvardy A, Mann C. Nature (London) 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 9.Harding C V, France J, Song R, Farah J M, Chatterjee S, Iqbal M, Siman R. J Immunol. 1995;22:1767–1775. [PubMed] [Google Scholar]
- 10.Palombella V J, Rando O J, Goldberg A L, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 11.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 12.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S. Science. 1995;268:726–730. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 13.Palmer J T. J Med Chem. 1995;38:3193–3196. doi: 10.1021/jm00017a002. [DOI] [PubMed] [Google Scholar]
- 14.Rohrwild M, Coux O, Huang H-C, Moerschell R P, Yoo S J, Seol J h, Chung C H, Goldberg A L. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupas A, Zwickl P, Baumeister W. Trends Biochem Sci. 1994;19:533–534. doi: 10.1016/0968-0004(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 16.Chuang S-E, Burland V, Plunkett G, Daniels D L, Blattner F R. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 17.Salter R D, Cresswell P. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nandi D, Iyer M N, Monaco J J. Exp Clin Immunogenet. 1996;13:20–29. [PubMed] [Google Scholar]
- 19.Riese R J, Wolf P R, Brömme D, Natkin L R, Villadangos J A, Ploegh H L, Chapman H A. Immunity. 1996;4:357–366. doi: 10.1016/s1074-7613(00)80249-6. [DOI] [PubMed] [Google Scholar]
- 20.Wiertz E J H J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 21.Weirtz E J H J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Nature (London) 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 22.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 23.Craiu, A., Gaczynska, M., Akopain, T., Gramm, C. F., Fenteany, G., Goldberg, A. L. & Rock, K. L. (1997) J. Biol. Chem., in press. [DOI] [PubMed]
- 24.Briane D, Olink-Coux M, Vassy J, Oudar O, Huesca M, Scherrer K, Foucrier J. Eur J Cell Biol. 1992;57:30–39. [PubMed] [Google Scholar]
- 25.Beersma M F C, Bijlmakers M J E, Ploegh H L. J Immunol. 1993;151:4455–4464. [PubMed] [Google Scholar]
- 26.Gaczynska M, Rock K L, Spies T, Goldberg A L. Proc Natl Acad Sci USA. 1994;91:9213–9217. doi: 10.1073/pnas.91.20.9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mozdzanowski J, Speicher D, Best S. Two-Dimensional Gel Electrophoresis. New York: Wiley; 1995. [Google Scholar]
- 28.Monaco J J, McDevitt H O. Proc Natl Acad Sci USA. 1982;79:3001–3005. doi: 10.1073/pnas.79.9.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]