Abstract
Rational protein design is an emerging approach for testing general theories of protein chemistry through the creation of new structures and functions. Here we present the first successful introduction by rational design of a [Fe4S4] cuboidal cluster into the hydrophobic core of Escherichia coli thioredoxin, a protein normally devoid of metal centers. Cuboidal [Fe4S4] is one of the stable forms of self-assembled iron–sulfur clusters that are thought to represent some of the earliest evolved biological redox centers. [Fe4S4] clusters have been recruited for use in a variety of proteins whose functions are central to many of the major biochemical processes ranging from simple soluble electron-transfer agents, to membrane-bound components of electron-transfer chains, to electron reservoirs in complex metalloenzymes such as nitrogenase. By situating an [Fe4S4] cluster into a protein environment not previously adapted by evolution we can explore the factors by which their activity is modulated by the protein matrix.
Keywords: rational protein design, FeS cluster environment, oxidation reduction, high-potential FeS cluster
Metalloproteins offer a particularly interesting target for the design of function, because the biological chemistry of metals is extraordinarily rich. Cuboidal [Fe4S4] clusters are among the most common electron-transfer centers found in biology (1). These clusters act as either simple soluble electron-transfer agents, membrane-bound components of electron-transfer chains, or parts of the electron reservoir found in complex metalloenzymes in plants, animals, and bacteria. In addition to being intimately involved in electron-transport systems and in the metabolism of carbon, oxygen, hydrogen, sulfur, and nitrogen, studies have suggested that these centers also exhibit gene regulatory, catalytic, and structural functions, and have been found as part of a morphogenetic protein (1).
The successful construction of a functional metal center requires that three sets of factors are taken into account: (i) the construction of a correctly folded protein, (ii) the coordination requirements of the metal, (iii) the modulation of the properties of the metal center and protein matrix to achieve the required control of reactivity. Thus far, metalloprotein designs have focused primarily on the first two factors (2–8). The redox properties of [Fe4S4] centers, which are strongly dependent on the protein environment, offer a good system to explore the modulation of the metal center by the protein (1, 9). Cuboidal [Fe4S4] proteins, which contain structurally equivalent metal clusters, are grouped in two classes, the ferredoxins (Fds) and the high-potential iron–sulfur proteins (HiPIPs). While the inorganic cluster can accommodate an overall charge of [Fe4S4]3+/2+/+, only one redox couple is accessible under physiological conditions; Fds use the [Fe4S4]+2/+1 couple (Eo = +80 mV to −700 mV vs. NHE), HiPIPs use the [Fe4S4]+3/+2 couple (Eo = +100 mV to +450 mV). Although several high-resolution x-ray crystallographic structures of HiPIP systems as well as 4Fe-, 7Fe-, and 8Fe-Fds are available (1), the identification of those factors that determine the redox couple are just beginning to emerge. Conjectures have been presented that cluster–protein (amide N—H to cluster S2− or cysteinyl S) hydrogen bonding (10), solvent accessibility of cluster, and electrostatic field gradients arising from the presence of acidic, basic, and hydrophobic residues in the vicinity of the [Fe4S4] cluster, are important in determining the redox potential (9). Spectroscopic studies (11), ab inito calculations (12), and computer simulations (13, 14) support these factors as determinants of cluster redox potential. Studies of model systems (15), mutant iron–sulfur proteins (16, 17), and interspecies variants (18) have also indicated the importance of the protein matrix and solvent. In addition, recent quantitative modeling simulations of the redox potentials of iron–sulfur proteins using the protein dipoles Langevin dipoles model have implicated the Coulombic interaction of the cluster with the protein atom charges as a major determinant of the variations in measured redox potentials (9). Specifically, the proximity and orientation of protein matrix dipoles arising from the parallel alignment of trans amide N—H and C=O bonds, which depends on the intrinsic structure of the polypeptide chain in the vicinity of the cluster, leads to substantial cluster charge–amide dipole interactions that influence the redox potential. These theories cannot be tested directly in natural proteins because that would require mutations of the backbone that cannot be achieved by normal means.
The rational design approach adopted here is to build a metal center into a protein frame of known structure, which normally does not possess such a center. The rational protein design algorithm DEZYMER (19) is used to search the three-dimensional structure of the host protein thioredoxin (Trx) to identify locations where it is predicted to be geometrically possible to introduce mutations to form the correct primary coordination sphere. Mutations are chosen to satisfy both the intended metal binding geometry and the steric requirements of the protein fold. Once a coordination sphere has been positioned, further mutations may be introduced to retain steric compatibility between the metal binding site and the surrounding protein. All the substitutions are therefore structurally conservative in nature, the intent being to maintain the original fold and stability of the host protein. Recently, we have successfully used this approach to design and construct a catalytically active iron superoxide dismutase site in the interior of Trx (44). Here we demonstrate how a HiPIP cluster can be constructed by placing a cuboidal [Fe4S4] cluster in the hydrophobic interior of the protein, Trx (Trx-Fe4S4).
MATERIALS AND METHODS
Molecular Simulations.
The DEZYMER (19) algorithm was used to perform the calculations for the site design and refinement on a NeXT computer.
Cloning and Protein Expression.
Trx-Fe4S4 was constructed by Kunkel mutagenesis techniques and the modified gene was inserted into the expression vector pKK-T7E (20) downstream of a T7 promoter and sequenced in full. Expression of Trx-Fe4S4 was achieved in Escherichia coli BL21(DE3). pTrx-Fe4S4/BL21 was grown in 2xYT/50 μg/ml ampicillin at 25°C and induced with isopropyl β-d-thiogalactoside; cells were harvested 4 hr after induction.
Purification of apo Trx-Fe4S4.
The harvested cells from 10 liters of culture were resuspended in 140 ml of 20 mM Tris⋅Cl (pH 7.5), 100 mM NaCl, 10 mM EDTA, and lysed by sonication. The cleared lysate was brought to 0.25% polyethyleneimine, centrifuged, and then brought to 45% saturation with solid ammonium sulfate and clarified. The supernatant was then brought to 90% saturation with solid ammonium sulfate. After centrifugation, the pellet was dissolved in 50 ml of 50 mM Tris⋅Cl (pH 8.1)/20 mM NaCl and dialyzed against the same buffer for 4 hr. The volume was reduced by dialysis against solid polyethylene glycol (PEG) 8000 at room temperature for 3 hr, followed by dialysis against 50 mM Tris⋅Cl (pH 8.1)/20 mM NaCl for 6 hr.
The sample was then applied in approximately 60 1.2-ml portions onto a 16 mm × 60 cm S75 gel filtration column and eluted with 50 mM Tris⋅Cl (pH 8.1)/20 mM NaCl at 4°C. Peak fractions were then combined and loaded onto a 25 mm × 7 cm DEAE-cellulose column, washed with five column volumes of 50 mM Tris⋅Cl (pH 8.1)/20 mM NaCl, and eluted with a 20–500 mM salt gradient in 20 mM Tris⋅Cl (pH 8.1). The isolated protein was concentrated 4-fold in an ultrafiltration cell and an equal volume of glycerol was added. Solutions of purified apo Trx-Fe4S4 were made 100 mM in DTT prior to storage at −70°C. This purification procedure resulted in the isolation of approximately 600 mg of apo Trx-Fe4S4 of >99% purity from a single 10 liter fermentor growth.
Cluster Reconstitution of Trx-Fe4S4.
Reconstitution of apo Trx-Fe4S4 is performed under strict anaerobic conditions in an inert (argon) atmosphere box using rigorously deoxygenated solutions. Apo Trx-Fe4S4 is passed through a gel filtration column eluted with 15 mM 2-(N-cyclohexylamino)ethanesulfonic acid (Ches), pH 8.5, to remove glycerol and DTT. The resulting solution is then made 2.0 M urea and 10 mM β-mercaptoethanol (βME) and allowed to sit for 15 min at 25°C. A freshly prepared solution of the synthetic cluster [Fe4S4(S-EtOH)4](Me4N)2 (21) in dimethyl sulfoxide (DMSO) is added to the urea-treated apo Trx-Fe4S4 protein in a 1:1 (cluster:protein) molar ratio, keeping the DMSO content of the protein containing solution <1.5%. After 5 min, the solution is diluted with an equal volume of 15 mM Ches (pH 8.5) solution. Excess FeS, βME, and residual urea are removed by desalting on a PD10 column equilibrated in 10 mM 4-morpholinepropane sulfonic acid (Mops) (pH 7.4)/100 mM NaCl. Reconstitution occurs with >90% recovery of protein. Fe:S2−:protein ratios are determined by atomic absorption spectroscopy (Varian SpectrAA20), colorimetric (22), and UV (reduced apo Trx-Fe4S4: ɛM = 13,700 verified by total amino acid analysis, Keck Foundation Biotechnology Laboratory, Yale University) spectrophotometric methods, respectively.
Measurement of in Vivo Activity of Trx.
An M13mp19 recombinant phage containing the gene for Trx-Fe4S4 was plated on a bacterial host strain deleted for wild-type Trx [A307 (23, 24)]. This strain will not support growth of wild-type M13mp19 phage, but will support the growth of an M13mp19 recombinant phage into which an active Trx has been cloned. Activity was scored by comparing phage titers on the ΔtrxA (A307) strain and its parent trxA+ strain, K38, of phage stock that had been grown on the permissive strain DH5αF′: a recombinant phage titer within one order of magnitude on both strains (typically 1010 to 1011 particles per ml) was scored as displaying Trx activity (inactive recombinants give titers of 105 to 106 particles per ml).
Spectroscopic Studies.
UV/vis electronic spectra were recorded under strict anaerobic conditions in gas tight optical cuvettes (1 cm pathlength) with a Perkin–Elmer Lambda 6 spectrometer. Circular dichroism (CD) spectra were obtained with an AVIV model 60DS Circular Dichroism Spectropolarimeter using a quartz cell with a 1 mm pathlength at 25°C at a protein concentration of approximately 25 μM. EPR spectra were collected on a Varian E-line EPR spectrometer operating at a microwave frequency of 9.24 GHz, with 1.5 mW microwave power, 10 G modulation amplitude at 25 K.
RESULTS
Rational Design.
The [Fe4S4Cys4]n− site definition was derived from the x-ray crystal structure of oxidized Chromatium vinosum HiPIP (1HIP) (25). However, since cluster structures vary little between different proteins and core oxidation states (26), our template, which includes all possible cysteine rotamers to allow maximum flexibility, should be considered as a generalized [Fe4S4(Cys)4]n− core. The DEZYMER algorithm generated several solutions for the [Fe4S4Cys4]n− site definition; we report the construction and characterization of one solution (Trx-Fe4S4; Leu-24-Cys, Leu-42-Cys, Val-55-Cys, and Leu-99-Cys, Fig. 1A). In addition to the four mutations required to introduce the [Fe4S4] cluster binding residues, Cys-32-Ser and Cys-35-Ser mutations were made to remove the native Trx disulfide bond, thereby eliminating any interaction from cysteine residues that are not part of the designed site. An isosteric Asp-26-Leu mutation was also introduced to improve the global stability of the protein; the Asp-26-Ala mutant of Trx is stabilized by 3 kcal compared with the wild-type protein (27, 28). The [Fe4S4(Cys)4]n− design moiety represents an isovolume exchange; there is only a negligible decrease in the volumes occupied by amino acid side chains in the interior of engineered Trx-Fe4S4 relative to Trx (Fig. 1B).
Figure 1.
Ribbon and space-filling representations of the designed cuboidal iron–sulfur protein Trx-Fe4S4. Iron is cyan, sulfur yellow. (A) Ribbon diagram showing the location of all mutations used in the construction of Trx-Fe4S4 and the region of the Trx host protein implicated in phage assembly. The cuboidal [Fe4S4] cluster binding residues (Leu-24-Cys, Leu-42-Cys, Val-55-Cys, and Leu-99-Cys) are buried between the central β-sheet and two α-helices. Cys-32-Ser and Cys-35-Ser mutations were made to remove the native Trx disulfide bond, thereby eliminating any interaction from cysteine residues that are not part of the designed site. An isosteric Asp-26-Leu mutation was also introduced to improve the global stability of the protein; the Asp-26-Ala mutant of Trx is stabilized by 3 kcal compared with the wild-type protein (27, 28). The residues that are believed to be important for protein–protein interactions in Trx function (32–35, 75, 76, 91–93) are colored red (29). (B) Space-filling representation of residues 24, 42, 55, and 99 in native E. coli Trx (Left) and Trx-Fe4S4 (Right). Incorporation of the [Fe4S4(Cys)4]n− design moiety represents a conservative isovolume exchange; there is only a negligible decrease in the volumes occupied by amino acid side chains in the interior of the designed iron–sulfur protein relative to the host.
Trx, a relatively small (108 amino acid residues) monomeric protein that contains a single redox active disulfide bond (Cys-32, Cys-35) (30), was chosen (24) as our initial scaffold protein because the gene for Trx has been cloned, sequenced, and expressed at high levels (31). Trx is a stable protein (oxidized Trx Tm, 86°C; reduced Trx Tm, 75°C) (30) that accommodates conservative as well as some nonconservative mutations (32). In addition to the availability of a high-resolution (1.68 Å) x-ray structure (33), there are several solution structures (oxidized and reduced) derived from NMR spectroscopic methods (34, 35), indicating the feasibility of obtaining structural information on the designed protein in the absence of crystals.
Folding of Designed Protein.
Because the designed substitutions are conservative in nature, maintenance of the protein fold is one of the design aims. The presence of a near-native Trx fold can be tested in vivo using the absolute requirement of Trx for phage M13 growth (23, 24, 36). Studies have shown that the redox properties of Trx are not necessary as the double Cys-32-Ser, Cys-35-Ser mutant is capable of catalyzing phage assembly (23). Mutagenesis studies suggest that the region important for protein–protein interactions in Trx function is formed by residues Gly-33, Pro-34, Ile-75, Pro-76, Val-91, Gly-92. and Ala-93 (29). It was found that Trx-Fe4S4 supported a normal phage titer when expressed in a nonpermissive ΔTrx− E. coli strain, providing strong evidence that the designed protein adopts a native Trx conformation (29).
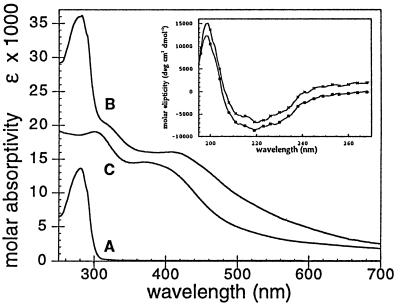
Further evidence for the proper folding of Trx-Fe4S4 is observed in the CD spectrum (195–270 nm) of purified apo Trx-Fe4S4 (Fig. 2 Inset), which is identical to that of native Trx. In addition, both apo Trx-Fe4S4 and native Trx have identical gel filtration retention times (fast protein liquid chromatography, 16 mm × 60 cm Superdex 75pg, Pharmacia), indicating equivalent Stokes radii. The combined phage assay results, CD spectrum, and gel filtration data strongly suggest that the overall aggregate structural elements of Trx are unaltered in the designed Trx-Fe4S4 protein.
Figure 2.
Optical spectrum of apo Trx-Fe4S4 (trace A), holo Trx-Fe4S4 (trace B) in 15 mM Mops/100 mM NaCl (pH 7.4) at 5°C, and the synthetic cluster [Fe4S4(S-EtOH)4](Me4N)2 (trace C) in 10 mM Ches/10 mM βME (pH 8.5). holo Trx-Fe4S4 contains 3.9 ± 0.2 mol of iron and 3.9 ± 0.2 mol acid labile sulfide per Trx protein; λmax = 280 nm, ɛM,280 = 36,000 λmax = 413 nm, ɛM,413 = 16,100. (Inset) CD spectra of apo Trx-Fe4S4 (upper trace) and Trx (lower trace) in 50 mM potassium phosphate (pH 7.0) at 25°C. The upper trace has been offset slightly to clarify the close superposition of the two curves.
Cluster Reconstitution of Purified Trx-Fe4S4.
As isolated, the overexpressed Trx-Fe4S4 contains no iron or elemental sulfur. Reconstitution of apo Trx-Fe4S4 was accomplished by exploiting the cooperative and reversible chemical denaturation process of the designed host protein (the midpoint of reduced apo Trx-Fe4S4 unfolding is 2.7 M urea vs. 4.6 M urea for reduced Trx) under strict anaerobic conditions in an argon flushed glove box at 25°C. The fully reduced protein was partially unfolded with 2.0 M urea in the presence of 10 mM mercaptoethanol and high pH (8.5) buffer. A synthetic, preformed tetranuclear iron–sulfur cluster (15, 21), [Fe4S4(S-EtOH)4](Me4N)2, dissolved in DMSO containing free βME (10 mM) was added to the unfolded protein in a 1:1 cluster:protein stoichiometric ratio. This soluble cluster is stable (<10%/hr loss) under these reaction conditions. Cluster incorporation is driven by ligand exchange processes where the less basic cysteine thiolate readily replaces the more basic mercaptoethanol ligand (26). Trx-Fe4S4 was then refolded by 1:1 dilution into the same high pH buffer, followed by removal of the remaining urea using gel filtration to exchange into 100 mM NaCl/15 mM Mops (pH 7.4) at 4°C. During this separation step, the cluster remained completely bound to the protein. Reconstituted Trx-Fe4S4 contains 3.9 ± 0.2 mol of iron (atomic absorption spectroscopy) and 3.9 ± 0.2 mol of acid labile sulfide (22) per mole of protein (reduced apo Trx-Fe4S4: ɛM = 13,700), consistent with the incorporation of a cuboidal [Fe4S4]n+ cluster. Gel filtration studies (10 mm × 35 cm Superdex 75, Pharmacia) of Trx-Fe4S4 show a single dominant species with the identical gel filtration retention time as wild-type Trx, indicating that holo Trx-Fe4S4 is a monomer and retains the global structure of Trx and apo Trx-Fe4S4.
Spectroscopic Properties of holo Trx-Fe4S4.
The UV/vis spectrum of holo Trx-Fe4S4 (Fig. 2) purified by gel filtration chromatography shows the broad visible absorbances (ɛM,413 = 16,100, based on total amino acid analysis) characteristic of [Fe4S4(Cys)4]2− proteins (1, 37). Based on SCF-Xα-SW theoretical models of the electronic structure of [Fe4S4(SR)4] clusters, the observed bands are proposed to arise from (RS−, S2−)-to-Fe charge transfer excitations (38). These studies suggest that the lower energy bands (420 nm), which show the most pronounced shifts (Δλmax ≈ 40 nm), involve excitation from orbitals with primarily S2− lone pair character, since these sites are less shielded from environmental effects than the thiolate sulfur centers. The smaller shift of the higher energy transition at 315 nm (Δλmax ≈ 12 nm) is thought to reflect differences in the basicity of the terminal mercaptoethanol and Cys thiolate ligands.
Fig. 2 also shows that the predicted perturbation to the electronic spectrum of the iron–sulfur cluster after its inclusion in the interior of the designed protein relative to the spectrum of the free synthetic cuboidal cluster is observed (15). A pronounced bathochromic shift of the iron–sulfur chromophore upon changing solvents from H2O to DMSO has been observed for both analogue clusters and iron–sulfur proteins and is believed to arise from a decrease in cluster solvent accessibility (15). This behavior is analogous to the shielding of a cluster initially in aqueous medium by a lower dielectric medium as would be expected for reconstitution of Trx-Fe4S4.
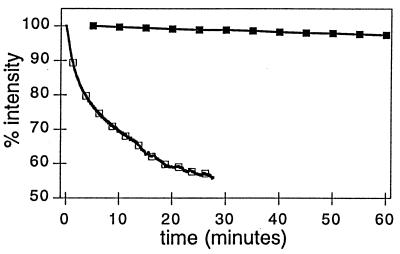
The stability of the protein-based chromophore (<5% decomposition hr−1, 4°C, argon atmosphere) is in direct contrast to the stability of the free synthetic cluster, which readily decomposes (t1/2 ≈ 3 min) in the absence of high concentrations of stabilizing exogenous thiol ligands (Fig. 3). These observations indicate that an intact cluster is sequestered in the interior of holo Trx-Fe4S4, thereby shielding it from hydrolytic cluster degradation pathways. These optical data of Trx-Fe4S4 are therefore consistent with a buried [Fe4S4] cuboidal cluster.
Figure 3.
Stability of the free synthetic cluster [Fe4S4(S-EtOH)4](Me4N)2 (□) in the presence of low levels (0.4 mM) of stabilizing exogenous βME and reconstituted holo Trx-Fe4S4 (▪) in the absence of stabilizing exogenous βME. Solution conditions: 10 mM Mops/100 mM NaCl (pH 7.4) at 5°C.
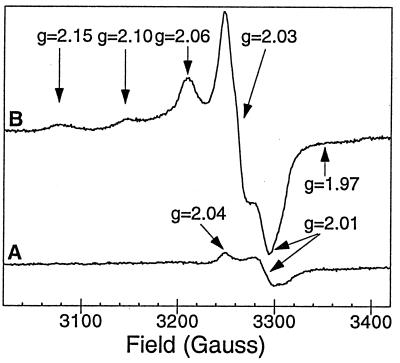
As isolated, reconstituted Trx-Fe4S4 is EPR silent, with only minor contributions (0.08 spin per protein) believed to arise from paramagnetic cluster contamination originating from [Fe4S4] cluster fragmentation. This resting state property is a direct consequence of the incorporation of a synthetic cluster consisting of [Fe2+Fe3+]2 in which one [Fe2+Fe3+] (S = 9/2) unit is antiferromagnetically coupled to a second [Fe2+Fe3+] (S = 9/2) to yield an S = 0 cluster (Fig. 4) (26). Upon oxidation of holo Trx-Fe4S4 with potassium ferricyanide (1.0 mM final concentration, ≈10 equivalents), an EPR-active species is formed whose spectral features are analogous to the [Fe4S4(Cys)4]− clusters found in oxidized HiPIPs. This species, which is observed over a wide temperature range (5 K to >40 K), consists of a dominant low field absorptive signal at g = 2.06, a major derivative signal at g = 2.03, and a high field signal at g = 2.01 (gavg ≈ 2.03). The low intensity signals at g = 2.15, 2.10, and 1.97, not observed in the diamagnetic Trx-Fe4S4 resting state, originate from the ferricyanide oxidation of the iron–sulfur protein. The major and minor groups of EPR features saturate together with P1/2 = 11.8 mW at 25 K and are indicative of transition metal-based unpaired electron density; signals typically integrate to 0.4 spin per protein.
Figure 4.
EPR spectrum of holo Trx-Fe4S4 in 15 mM Mops/100 mM NaCl (pH 7.4), protein concentration 70 ± 5 μM. (Trace B) holo Trx-Fe4S4 oxidized with 10 equivalents of K3[Fe(CN)6]. (Trace A) Untreated (resting state) holo Trx-Fe4S4. EPR spectra collected on a Varian E-line EPR spectrometer operating at 9.24 GHz, 1.5 mW observe power, 10 G modulation amplitude, 25 K.
The EPR properties of Trx-Fe4S4 are analogous to those reported for HiPIPs in that both are EPR active only upon oxidation, display signals with gavg >2, and exhibit similar microwave power and temperature dependencies (1). Although the observed g values of 2.06, 2.03, and 2.01 vary from those g values reported for natural HiPIP centers (1), we feel that this may be a consequence of the protein environment that surrounds the cluster or to subtle structural distortions of the [Fe4S4] cluster itself imposed by the designed protein host. Future studies examining the Mössbauer and magnetic CD properties of holo Trx-Fe4S4 will undoubtedly shed light on the electronic coupling within the sequestered cluster. Attempts to generate the reduced paramagnetic [Fe4S4(Cys)4]3− cluster with dithionite (≤100 equiv.) lead only to cluster degradation. This oxidation/reduction behavior is a characteristic of HiPIP iron–sulfur centers and distinguishes them from Fds and from other cluster types (39).
While HiPIP [Fe4S4(Cys)4]− clusters are typically characterized by axial EPR signals with gavg > 2, reduced FD [Fe4S4(Cys)4]3− clusters typically show rhombic EPR signals (g = 2.06, 1.92, 1.88) with gavg < 2 (1, 39). The EPR data in Fig. 4 similarly rule out a dominant [Fe3S4] cluster, whose oxidized ground state should contain three antiferromagnetically Fe3+ (S = 5/2) centers yielding an S = 1/2 ground state characterized by isotropic or axial EPR signals exhibiting gavg values between 2.02–1.96 with a low-field g value of 2.02 (1). As solvent accessible [Fe4S4(Cys)4]2− and [Fe4S4(S-EtOH)4]2− clusters are stable to reduction but not oxidation, and exhibit EPR signals with gavg < 2 (40), the spectral properties of Trx-Fe4S4 cannot be due to a cluster bound to the exterior of the protein, in agreement with the interpretation of the optical spectrum data.
DISCUSSION
We have used the rational design algorithm, DEZYMER, to introduce a cuboidal [Fe4S4] cluster into the hydrophobic interior of E. coli Trx, a protein normally devoid of transition metal centers. Inspection of the designed site reveals several favorable properties. Trx folds to form a core of β-pleated sheet flanked on either side by two α-helices; the structure can be considered as being formed of two domains, βαβαβα from residues 1 to 59 (domain 1) and ββα from residues 76 to 108 (domain 2) (33). The DEZYMER solution (Fig. 1), which is located between the central β-sheet (Cys-24, β-strand 2; Cys-55, β-strand 3) and one α-helix (Cys-42, α-helix 2; Cys-99, α-helix 4) from each domain, is buried approximately 8–9 Å from the surface. All peptide amide NH groups in the vicinity of the designed site participate in secondary structure interactions in this region and are unavailable for hydrogen bonding to the cluster in the model. Finally, the cluster site is buried in the interior of the Trx host and surrounded by a hydrophobic shell generated by Ile-41, Ile-45, Ala-46, Leu-53, Leu-78, Leu-94, and Leu-103.
The DEZYMER algorithm is based on the premise that the mutations introduced are conservative and maintain the overall protein fold of the host scaffold (inverse folding constraint). This assumption was tested by comparing the in vivo biological properties of Trx-Fe4S4 and wild-type Trx. The cluster binding site was introduced into a region of Trx that is located near the putative protein–protein interaction surface required for the growth of filamentous phages, thereby allowing a test of whether the mutations in Trx-Fe4S4 affect the original functions of Trx. Trx-Fe4S4 exhibited full activity, thereby providing strong evidence that the designed protein adopts a wild-type conformation or at least maintains the essential structural features of the Trx surface that are necessary for this in vivo activity. Furthermore, both the CD spectra (apo) and hydrodynamic (apo, holo) properties of Trx-Fe4S4 are equivalent to the Trx host, offering both genetic and physical data suggesting that the overall structure of Trx-Fe4S4 is essentially indistinguishable from Trx. The UVCD spectrum of holo Trx-Fe4S4 is dominated by signals from the iron–sulfur cluster and is therefore relatively uninformative regarding the protein fold. Complete structural determinations of apo and holo Trx-Fe4S4, which are ultimately required to assess the success of the design, are currently underway.
Evidence for cuboidal cluster incorporation into the designed site comes from both optical and EPR studies. The excellent stability of the Trx-Fe4S4 chromophore in the absence of free thiols, the bathochromic shift in the cluster chromophore upon protein reconstitution, coupled with an EPR silent resting state and the observation of a paramagnetic cluster upon cluster oxidation all indicate the presence of a protected cuboidal [Fe4S4] cluster. Determination of the expected Fe:S2−:Trx, the similarity of the Trx-Fe4S4 chromophore to that of known Fe4S4Cys4 proteins, and it’s dissimilarity to other (Fe2S2Cys4, FeCys4) cluster types argues against the possibility of cluster fragmentation accompanied by iron binding.
Ferricyanide oxidation also places a limit on the redox properties of the cluster, consistent with its HiPIP-like properties. The oxidized Trx-Fe4S4 cluster is only transiently stable, as evidenced by rapid bleaching of the visible absorbance upon ferricyanide addition, and the disappearance of the gavg > 2 EPR signal upon prolonged incubation at 4°C. Such cluster degradation has been reported for HiPIPs and is believed to produce reducing equivalents (Fe2+, S2−, and CysSH) (41). While these factors complicate the determination of a redox potential, our observation of complete oxidation upon addition of >7 equivalents of potassium ferricyanide, and incomplete oxidation at lower concentrations allows us to place a lower bound of +300 mV vs. NHE on the redox potential of Trx-Fe4S4, based on the Nernst equation and the [ferricyanide]/[ferrocyanide] ratio. This potential is within the range of redox potentials observed for HiPIPs (Eo = +100 mV to +450 mV).
Thus, the key aspects of our design have been realized; (i) we have successfully created a cluster of the correct topological type, (ii) holo Trx-Fe4S4 maintains the folded structure of its unmodified host protein, (iii) incorporation of a cuboidal [Fe4S4(Cys)4]2−/1− cluster in the hydrophobic interior of Trx is sufficient to generate a HiPIP-like metalloprotein. The DEZYMER algorithm makes predictions based on simple geometric principles without explicit considerations of binding thermodynamics or protein dynamics. We have demonstrated here that these approximations can have sufficient predictive power to redesign the hydrophobic interior of a protein and introduce a [Fe4S4] cluster.
The designed site does not follow any of the consensus sequences observed in proteins containing cuboidal [Fe4S4] clusters (Table 1) (1). The consensus sequence for coordination of a [Fe4S4] cluster in bacterial Fds is Cys-X2-Cys-X2-Cys with a more remote Cys residue supplying the fourth ligating sulfur center. Similarly, in 8Fe Fds, the consensus sequence involves two Cys-X2-Cys-X2-Cys-X3-Cys-Pro sequences with each cluster coordinated by three Cys residues from one group and a more remote Cys residue. Studies have demonstrated the successful construction of a Fd-like clusters from synthetic polypeptides derived from the consensus sequence of natural Fds (42, 43). Different arrangements of coordinated Cys residues are also found for the Fe4S4 clusters of HiPIP proteins (Cys-X2-Cys-X16-Cys-X13-Cys). The coordinating Cys residues in Trx-Fe4S4 are found in the pattern Cys-X18-Cys-X13-Cys-X44-Cys, which does not follow the standard Fd or HiPIP patterns for creating the ligand sphere for the [Fe4S4] cluster. The existence of the Cys residues at locations widely separated in primary sequence but appropriately positioned in the three-dimensional space of the folded scaffold protein offers the unique opportunity to begin to probe the effect of placing these clusters in different secondary structural environments, allowing separation of fundamental chemical requirements from the accidents of history imposed by evolution.
Table 1.
Identified consensus sequences for cuboidal [Fe4S4] clusters (1)
| ---Cys-X2-Cys-X2-Cys-----Cys-Pro--- | 4Fe Fd |
| ---Cys-X2-Cys-X2-Cys-X3-Cys-Pro-----Cys-X2-Cys-X2-Cys-X3-Cys-Pro--- | 8Fe Fd |
| ---Cys-X2-Cys-X2-Cys-X3-Cys-Pro-----Cys-X2-Cys-X7,8-Cys-X3-Cys-Pro--- | 8Fe Fd |
| ---Cys-X7-Cys-X3-Cys-Pro-----Cys-X2-Cys-X2-Cys-X3-Cys-Pro--- | 7Fe Fd |
| ---Cys-X6-Cys--- ---Cys-X6-Cys--- | 4Fe in PSI |
| ---Cys-X34-Cys--- ---Cys-X34-Cys--- | N2ase Fe protein |
| ---Cys-----Cys-X2-Cys--- | Aconitase |
| ---Cys-X6-Cys-X2-Cys-X5-Cys--- | Endonuclease III |
| ---Cys-X2-Cys-X16-Cys-X13-Cys--- | HiPIP |
| ---Cys-X18-Cys-X13-Cys-X44-Cys--- | Trx-Fe4S4 |
Recent simulation studies have shown that correlating variations of redox potential with protein environment requires assessing the Coulombic interaction of the cluster with the protein (the major determinant), the polarizability of the protein, and the interaction of the cluster and protein with solvent (9). These results suggest that the folding of the polypeptide in the vicinity of the cluster (proximity and orientation of cluster charge–amide dipole interaction) is a major factor in fixing the redox potential; side chain groups are proposed to be of secondary importance. The DEZYMER algorithm places no a priori constraint on the choice of the secondary structural environment in which the new place is incorporated, offering the opportunity to test this hypothesis. The fact that the orientation of the amide dipoles in the model of Trx-Fe4S4 are fixed by the folded structure of Trx and not optimized for the presence of the incorporated cluster is consistent with the HiPIP-like redox properties of Trx-Fe4S4 (9). Finally, the geometry of the designed cluster is such that neighboring hydrophobic side chains can be mutated to introduce hydrogen bonding or charged residues to systematically probe the role of these determinants in the electronic properties and redox potential of the cluster.
Acknowledgments
We greatly appreciate the efforts and insights of Ann L. Pinto during these investigations. We also thank Gary Brudvig and Veronika Szali for assistance with EPR instrumentation. This work was supported in part by National Science Foundation Grant CHE-9419178 (J.P.C.), the Exxon Education Foundation (J.P.C.), the Alfred P. Sloan Foundation (J.P.C.), and National Institutes of Health Grant R29GM49871 (H.W.H.).
ABBREVIATIONS
- Fd
ferredoxin
- HiPIP
high-potential iron–sulfur protein
- Trx
thioredoxin
- DMSO
dimethyl sulfoxide
- βME
β-mercaptoethanol
- Mops
4-morpholinepropane sulfonic acid
- CD
circular dichroism
References
- 1.Johnson M K. In: Iron–Sulfur Proteins. King R B, editor. Vol. 4. Oxford: Wiley; 1994. pp. 1896–1915. [Google Scholar]
- 2.Bryson J W, Betz S F, Lu H S, Suich D J, Zhou H X, O’Neil K T, DeGrado W F. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 3.Higaki J N, Fletterick R J, Craik C S. Trends Biochem Sci. 1992;17:100–104. doi: 10.1016/0968-0004(92)90245-5. [DOI] [PubMed] [Google Scholar]
- 4.Matthews D J. Curr Opin Biotechnol. 1995;6:419–424. doi: 10.1016/0958-1669(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 5.Regan L. Trends Biochem Sci. 1995;20:280–285. doi: 10.1016/s0968-0004(00)89044-1. [DOI] [PubMed] [Google Scholar]
- 6.Tawfik D S, Eshhar Z, Green B S. Mol Biotechnol. 1994;1:87–103. doi: 10.1007/BF02821512. [DOI] [PubMed] [Google Scholar]
- 7.Hellinga H W. Curr Opin Biotechnol. 1996;7:437–441. doi: 10.1016/s0958-1669(96)80121-2. [DOI] [PubMed] [Google Scholar]
- 8.Hellinga H W. In: Protein Engineering: Principles and Practice. Cleland J L, Craik C S, editors. New York: Wiley–Liss; 1996. pp. 369–398. [Google Scholar]
- 9.Stephens P J, Jollie D R, Warshel A. Chem Rev. 1996;96:2491–2513. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 10.Adman E T, Watenpaugh K D, Jensen L H. Proc Natl Acad Sci USA. 1975;72:4854–4858. doi: 10.1073/pnas.72.12.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backes G, Mino Y, Loehr T M, Meyer T E, Cusanovich M A, Sweeney W V, Adman E T, Sanders-Loehr J. J Am Chem Soc. 1991;113:2055–2064. [Google Scholar]
- 12.Sheridan R P, Allen L C, Carter C W. J Biol Chem. 1981;256:5052–5057. [PubMed] [Google Scholar]
- 13.Jensen G M, Warshel A, Stephens P J. Biochemistry. 1994;33:10911–10924. doi: 10.1021/bi00202a010. [DOI] [PubMed] [Google Scholar]
- 14.Langen R, Jensen G M, Jacob U, Stephens P J, Warshel A. J Biol Chem. 1992;267:25625–25627. [PubMed] [Google Scholar]
- 15.Hill C L, Renaud J, Holm R H, Mortenson L E. J Am Chem Soc. 1977;99:2549–2557. doi: 10.1021/ja00450a024. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal A, Li D, Cowan J A. Proc Natl Acad Sci USA. 1995;92:9440–9444. doi: 10.1073/pnas.92.21.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwagami S G, Creagh A L, Haynes C A, Borsari M, Felli I C, Piccioli M, Eltis L D. Protein Sci. 1995;4:2562–2572. doi: 10.1002/pro.5560041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heering H A, Bulsink B M, Hagen W R, Meyer T E. Biochemistry. 1995;34:14675–14686. doi: 10.1021/bi00045a008. [DOI] [PubMed] [Google Scholar]
- 19.Hellinga H W, Richards F M. J Mol Biol. 1991;222:763–786. doi: 10.1016/0022-2836(91)90510-d. [DOI] [PubMed] [Google Scholar]
- 20.Kriwacki R W, Schultz S C, Steitz T A, Caradonna J P. Proc Natl Acad Sci USA. 1992;89:9759–9763. doi: 10.1073/pnas.89.20.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christou, G. & Garner, C. D. (1979) J. Chem. Soc. Dalton Trans, 1093–1094.
- 22.Beinert H. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 23.Russel M, Model P. J Biol Chem. 1986;261:14997–15005. [PubMed] [Google Scholar]
- 24.Hellinga W H, Caradonna J P, Richards F M. J Mol Biol. 1991;222:787–803. doi: 10.1016/0022-2836(91)90511-4. [DOI] [PubMed] [Google Scholar]
- 25.Carter C W J, Kraut J, Freer S T, Alden R A, Sieker L C, Adman E, Jensen L H. Proc Natl Acad Sci USA. 1972;69:3526–3529. doi: 10.1073/pnas.69.12.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg J M, Holm R H. In: Iron–Sulfur Proteins. Spiro T G, editor. Vol. 4. New York: Wiley–Interscience; 1982. pp. 1–66. [Google Scholar]
- 27.Langsetmo K, Fuchs J A, Woodward C. Biochemistry. 1991;30:7603–7609. doi: 10.1021/bi00244a032. [DOI] [PubMed] [Google Scholar]
- 28.Langsetmo K, Fuchs J A, Woodward C, Sharp K A. Biochemistry. 1991;30:7609–7614. doi: 10.1021/bi00244a033. [DOI] [PubMed] [Google Scholar]
- 29.Eklund H, Cambillau C, Sjöberg B-M, Holmgren A, Jörnvall H, Höög J-O, Bränden C-I. EMBO J. 1984;3:1443–1449. doi: 10.1002/j.1460-2075.1984.tb01994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 31.Wallace B J, Kushner S R. Gene. 1984;32:399–401. doi: 10.1016/0378-1119(84)90015-5. [DOI] [PubMed] [Google Scholar]
- 32.Wynn R, Richards F M. Protein Sci. 1993;2:395–403. doi: 10.1002/pro.5560020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katti S K, LeMaster D M, Eklund H. J Mol Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 34.Dyson H J, Holmgren A, Wright P E. Biochemistry. 1989;28:7074–7078. doi: 10.1021/bi00443a044. [DOI] [PubMed] [Google Scholar]
- 35.Dyson H J, Gippert G P, Case D A, Holmgren A, Wright P E. Biochemistry. 1990;29:4129–4136. doi: 10.1021/bi00469a016. [DOI] [PubMed] [Google Scholar]
- 36.Russel M, Model P. Proc Natl Acad Sci USA. 1985;82:29–33. doi: 10.1073/pnas.82.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiro T G. Iron–Sulfur Proteins. Vol. 4. New York: Wiley; 1982. p. 423. [Google Scholar]
- 38.Yang C Y, Johnson K H, Holm R H, Norman J G J. J Am Chem Soc. 1975;97:6596–6598. doi: 10.1021/ja00855a062. [DOI] [PubMed] [Google Scholar]
- 39.Orme-Johnson W H, Orme-Johnson N R. In: Iron–Sulfur Proteins: The Problem of Determining Cluster Type. Spiro T G, editor. Vol. 4. New York: Wiley–Interscience; 1982. pp. 67–96. [Google Scholar]
- 40.R′zaigui M, Hatchikian E C, Benlian D. Biochem Biophys Res Commun. 1980;92:1258–1265. doi: 10.1016/0006-291x(80)90422-2. [DOI] [PubMed] [Google Scholar]
- 41.Bian S, Hermann C F, Hille R, Cowan J A. Biochemistry. 1996;35:14544–14552. doi: 10.1021/bi961658l. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura A, Ueyama N. In: Encyclopedia of Inorganic Chemistry. King R B, editor. Vol. 4. New York: Wiley; 1994. pp. 1883–1896. [Google Scholar]
- 43.Gibney B R, Mulholland S E, Rabanal F, Dutton P L. Proc Natl Acad Sci USA. 1996;93:15041–15046. doi: 10.1073/pnas.93.26.15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto A L, Hellinga H W, Caradonna J P. Proc Natl Acad Sci USA. 1997;94:5562–5567. doi: 10.1073/pnas.94.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]