Abstract
The Fe2+ ion that specifically replaces Mg2+ in the active center of RNA polymerase generates reactive hydroxyl radicals that cause highly localized cleavage of polypeptide chains. Mapping of the cleavage sites revealed the overall architecture of the active center. Nine distinct sites, five in the β subunit and four in the β′ subunit of Escherichia coli RNA polymerase, all at or near highly conserved sequence motifs, are brought together in the enzyme’s ternary structure within the distance of ≈1 nm from the active center Me2+. These sites are located in at least six different domains of the subunits, reflecting modular organization of the active center.
During the past decade, x-ray crystallographic analysis of small, single-subunit polymerases, such as DNA polymerase I, HIV reverse transcriptase, and RNA polymerase of bacteriophage T7 revealed a common principal structure whereby a single protein domain contains a characteristic cleft where the enzymatic reaction of phosphodiester bond formation is believed to occur (1). A similar cleft has been observed in the low-resolution contours of large multisubunit cellular RNA polymerases (RNAPs) (2–4), leading to speculation that their active center resembles that of the smaller enzymes. The excess protein in large polymerases, which amounts to hundreds of thousands of dalton, is explained by complex regulatory requirements of the cell.
But how are the larger polymerases built? Two principal types of organization are possible. First, a single catalytic domain structurally analogous to a single-subunit polymerase may be surrounded by regulatory components. Alternatively, different structural domains may participate in the active center of the large enzymes, forming the cleft that resembles that of the single-domain polymerases only in shape but not in structural design. Lack of sequence conservation between large and small polymerase families argues for the latter possibility. Here we present direct structural evidence to the same effect using the recently described technique of Fe2+-mediated local protein cleavage (5).
MATERIALS AND METHODS
Histidine-tagged RNA polymerase was derivatized at Met-932 of the β′ subunit (V.M., E. Lukhtanov, A.G., and A.M., unpublished data) and Lys-1242 (M.K., K. Severinov, A.G., and A.M., unpublished data) of the β subunit as described (5, 6), except that NaBH4 was added after [α-32P]UTP. Fe2+ induced autocleavage, and degradation of RNAP polypeptides at Met and Cys were performed as in (5). Products were analyzed by SDS/PAGE and gel slabs were autoradiographed as described (5). Gel concentrations are indicated in the figure legends.
RESULTS AND DISCUSSION
Fe2+-Mediated Cleavage of RNAP.
The Fe2+ ion in appropriate ligation under aerobic conditions generates hydroxyl radicals. These activated oxygen species are extremely reactive and cause degradation of biopolymers with a diffusion limited rate, within the range of ≈1 nm. The process is known as Fenton reaction (7) and is used for footprinting of nucleic acid–protein complexes. As was demonstrated previously (5), Fe2+ can bind to Escherichia coli RNA polymerase active center at the site normally occupied by Mg2+. The binding of Fe2+ leads to cleavage of both protein and DNA in the RNAP–promoter complex. The DNA is cleaved just upstream of the transcription start site, while the major protein cleavage occurs in the largest RNAP subunit, β′, near three evolutionarily conserved Asp residues that coordinate metal in the active center. In addition, several minor Fe2+-induced cleavages occur in the β and β′ subunits.
Here, the minor cleavages were mapped with the objective of deducing the overall architecture of the RNAP active center. To facilitate mapping, the β and β′ subunits were tagged with a radiolabeled reporter group near their C termini. As the tags, chimerical rifampicin–nucleotide compounds were used (6) that were synthesized and crosslinked to RNAP in open promoter complexes. Two different tagging protocols were employed, resulting in the addition of Rif-A*pU to Lys-1242 of the β subunit, and Rif-GpCpT* to Met-932 of the β′ subunit (the bold print symbolizes radioactive phosphate, the asterisk, the crosslinked nucleotide). After crosslinking, the enzyme was subjected to Fe2+ induced cleavage and the degradation products were analyzed by SDS/PAGE.
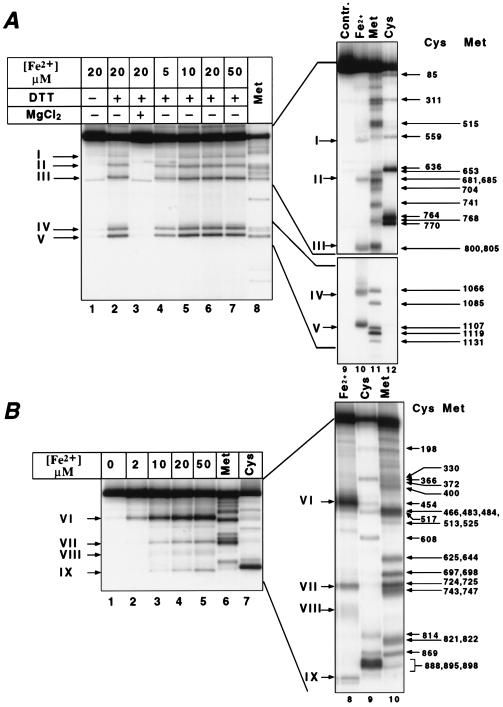
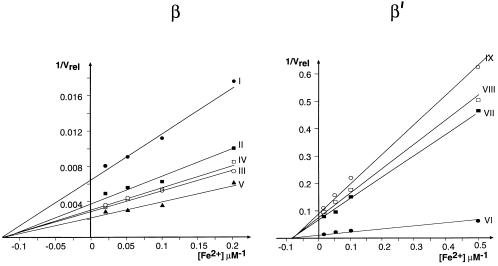
Five distinct cleavage sites could be seen in β (Fig. 1A), and four in β′ (Fig. 1B). In the absence of a reducing agent, the cleavage was weak (Fig. 1A, lane 1) in accordance with the free radical mechanism. No cleavage occurred in the presence of magnesium, which reflects the competitive nature of Fe2+ binding (Fig. 1A, lane 3). The efficiency of cleavage increased with the concentration of Fe2+ (Fig. 1A, lanes 4–7; Fig. 1B, lanes 2–5) and eventually reached a plateau reflecting the saturation of the binding site(s). Within the same subunit, different sites displayed the same dependence of cleavage rate on Fe2+ concentration. However, two different kinetics were observed for the two subunits. The apparent binding constants for Fe2+ calculated from the abscissa intercepts of the double reciprocal plots (Fig. 2) were ≈8 μM and ≈12 μM for β and β′, respectively.
Figure 1.
Fe2+-induced autocleavage of RNAP radiolabeled in the β (A) or β′ subunit (B). Histidine-tagged RNA polymerase was derivatized with radioactive reporter group at Met-932 of the β′ subunit, and Lys-1242 of the β subunit, and subjected to Fe2+ induced autocleavage (A, lanes 1–7 and 10; and B, lanes 2–5 and 8). Products were analyzed by SDS/PAGE followed by autoradiography. As size markers, products of chemical degradation at Met (A, lanes 8 and 11; B, lanes 6 and 10), or Cys (A, lane 12; B, lanes 7 and 9) was fractionated is the same gels. The following gel systems were used (acrylamide concentration/length): A Left, 8–16%/20 cm; A Right Upper, 8%/40 cm; A Right Lower, 12%/40 cm; B Left, 6%/20 cm; B Right, 6%/40 cm. The positions of Cys and Met residues in the sequence ladders are marked by arrows with Arabic symbols. Cleavage products are marked by arrows in Roman symbols.
Figure 2.
Dependence of cleavage rate on Fe2+ concentration in a double-reciprocal plot. Roman numbers indicate the products as in Fig. 1. Cleavage rate (Vrel) is expressed in arbitrary units derived from counting radioactive bands in wet gels using a Packard Instant Imager. The results and the apparent binding constant values (see text) were reproducible in three independent experiments.
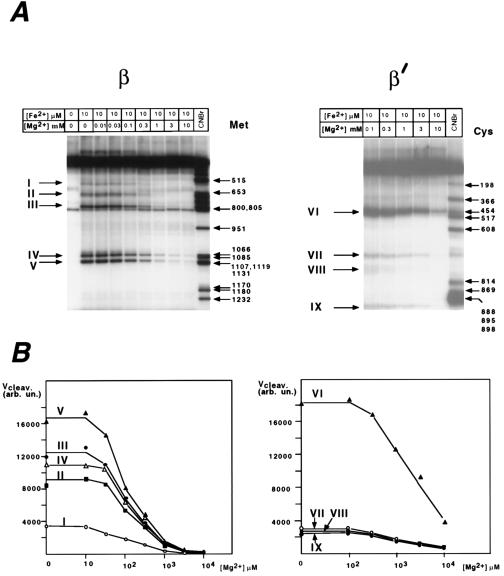
Fig. 3 shows the effect of increasing Mg2+ concentration on the rate of cleavage. Again, the protective effect of Mg2+ was the same for all of the cleavage sites within each subunit, whereas higher Mg2+ concentrations were required for the protection in the case of β′ than β.
Figure 3.
The protective effect of Mg2+ on Fe2+-induced cleavage. Roman numbers indicate the cleavage products as in Fig. 1. (A) Autoradiograms of gel slabs. (B) Radioactivity counts in product bands expressed in arbitrary units. The reaction conditions were as in Fig. 1, with varying concentrations of Mg2+ added.
The difference in the apparent affinity to Me2+ ions between the sites in β and β′ can in principle have two explanations. First, there may be two different Me2+ binding sites in the two subunits. Second, the apparent difference may be an artifact caused by different charge effect near the catalytic center introduced by the two reporter groups. We favor the latter explanation because the modification of β–Lys-1242 leaves intact the charge of lysine residue, whereas the modification of β′–Met-932 adds an extra positive charge. Moreover, β′–Met-932 is located closer to the catalytic center than the β′–Lys-1242 because of the 1-nucleotide difference in the position of the crosslink in the probes (Rif-GpCpT* vs. Rif-A*pU). The view that cleavage of β and β′ is caused by Me2+ binding in the same site is strongly supported by the fact that the DDD mutation abolished cleavage of both subunits (5).
Additional support to this view comes from the result, obtained for Mn2+ by kinetic and magnetic resionance methods, that there is only one tight binding site with less than 10 μM dissociation constant whereas other Me2+ sites in RNAP are ≈100 weaker (8). The constants for Fe2+ binding determined in this work is in the 10 μM range (see above). Thus we believe that in the concentrations range used, Fe2+ binding at the weak sites would be insignificant. Moreover, weak sites are weak because they poorly coordinate the Me2+ ion. Since coordination is essential for free radical generation, it is not likely that Fe2+, even if it was bound, would lead to cleavage at all.
Mapping of the Cleavage Sites.
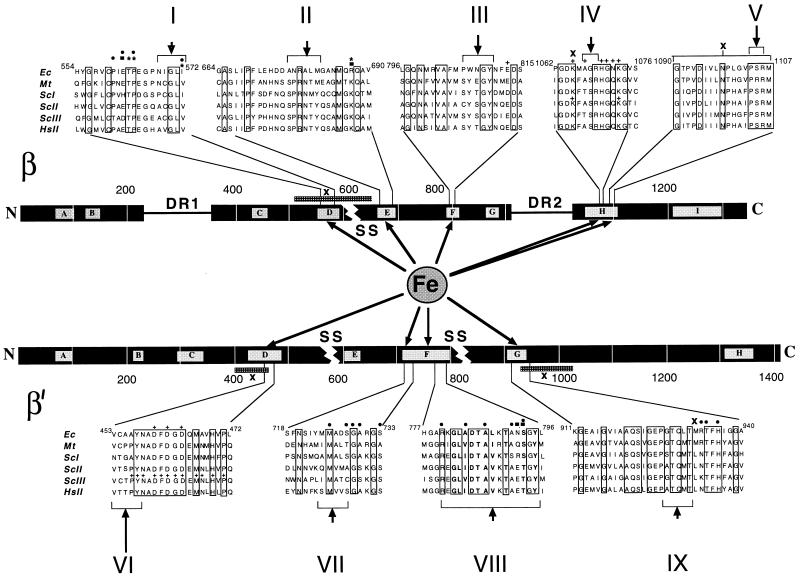
To decipher the architecture of the catalytic center we mapped the Fe2+ cleavage sites. To aid mapping, sequence markers were generated by chemical degradation of the radiotagged subunits at Cys and Met residues under the “single hit” conditions. Since the tags are situated close to a polypeptide terminus in both the β and the β′ subunits, single-hit degradation yields families of nested, easily identifiable fragments. The Fe2+ cleavage products along with the marker fragments were separated by SDS/PAGE and visualized by autoradiography (Fig. 1). From the relative migration of these products, the location of the cleavage sites could be discerned. In this way we mapped five Fe2+ cleavages in β, and four in β′ (Fig. 4). All of the cleavages occurred in highly evolutionarily conserved regions.
Figure 4.
Fe2+-induced cleavage sites in the β and β′ subunits. The subunits are represented by horizontal bars with the conserved regions symbolized by lettered boxes. Dispensable regions (DR1 and DR2) and chloroplast and archaebacterial “split sites” (SS) are indicated. Sequence alignments around the cleavage sites are as follows: Ec, E. coli; Mt, Methanobacterium thermoautotrophicum; ScI, ScII, and ScIII, Saccharomyces cerevisiae Pol I, Pol II, and Pol III, respectively; HsII, Homo sapiens Pol II. Invariant residues are boxed. Arrows point at Fe2+ induced cleavage sites; their extent is indicated by brackets. The map positions of known mutations are indicated as follows: ∗, Rifampicin resistant mutations in E. coli (9, 10); ⋅, termination-altered mutations in E. coli (12, 13); n, Streptolydigin resistant mutations in E. coli (11) and Bacillus subtilis (27); +, lethal substitutions in E. coli (15–17) and yeast (14, 18); x, crosslinking sites or segments (19–23).
For most sites, the accuracy of the mapping was from 2 to 6 amino acid residues with the exception of the β′-F region where the diffused appearance of the protein band (cleavage product VIII in Fig. 1B) apparently reflects multiple cleavages in this site. In fact, the relative intensity and band sharpness of the cleavage products may reflect the distance of the respective sites from the chelated Me2+ ion. Thus, the most prominent cleavage occurred in the β′-D region (product VI, Fig. 1B) where the active center Mg2+ is actually held (5).
Architecture of the Catalytic Center.
The topology of the active center that emerges from the Fe2+ cleavage data is in agreement with the results of previous mutational analysis of RNAP catalytic function (Fig. 4). The most severe mutation described is the triple alanine substitution DDD in the β′-D region which removes the aspartate residues coordinating Mg2+ in the active center (5). Most of other mutations have a subtler effect and have been interpreted as partial distortions of the catalytic center of varying severity (9–13, 15–17). Our present results indicate that all of these mutations distort the catalytic pocket directly, rather than from a distance via a conformational change. Our results are also in agreement with the previous data on affinity crosslinking, obtained with probes incorporated either in the priming nucleoside triphosphate (19–21) or in the 3′ terminus of nascent RNA (22, 23). These experiments have implicated β-D, β-H, β′-D, and β′-G regions in the formation of the catalytic pocket (Fig. 4).
Recent analyses of domain organization of RNAP subunits has shown that both β and β′ are built of distinct modules that need not be physically connected to fold and assemble into the functional RNAP molecule (24, 25). These modules correspond to evolutionary demarcations within the subunits evident from sequence alignments, such as nonconserved “dispensable regions” or “split sites” reflecting interruptions in the homologous sequences in chloroplasts and archaebacteria (Fig. 4). Checked against these demarcations, the sites participating in the active center uncovered in our work fall into at least six different modules. In the β subunit, these modules are separated by the archaebacterial split site between regions β-D and β-E, and by dispensable region II between regions β-G and β-H. In β′, regions β′-D and β′-E are separated by the chloroplast split site, and regions β′-F and β′-G, by the archaebacterial split site (Fig. 4).
Thus, the active center of the large cellular RNA polymerase fundamentally differs from that of small, single-subunit polymerases in basic architecture and evolutionary origin, even though they may share a common enzymatic mechanism and a similar orientation of active residues in the catalytic pocket. The modular organization of the active center may give the cellular enzyme greater flexibility needed for regulation. For example, one can imagine that a module participating in the active site at the same time serves as the target for an outside regulatory factor thus providing a direct physical “bridge” for conducting signals that modulate catalytic function. Also it is possible that relative movements of the modules constitute the basis for the apparent stretching of RNAP active center observed during early stages of elongation (26).
Acknowledgments
We are grateful to Sergei Borukhov and Evgeny Nudler for comments. This work was supported by Grant GM49242 and U.S./Russia Collaborative Grant TW00259 from the National Institutes of Health.
ABBREVIATION
- RNAP
RNA polymerase
References
- 1.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 2.Darst S A, Kubalek E W, Kornberg R D. Nature (London) 1989;340:730–732. doi: 10.1038/340730a0. [DOI] [PubMed] [Google Scholar]
- 3.Darst S A, Edwards A M, Kubalek E W, Kornberg R D. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 4.Schultz P, Celia H, Riva M, Sentenac A, Oudet P. EMBO J. 1993;12:2601–2607. doi: 10.1002/j.1460-2075.1993.tb05920.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 6.Mustaev A, Zaychikov E, Severinov K, Kashlev M, Polyakov A, Nikiforov V, Goldfarb A. Proc Natl Acad Sci USA. 1994;91:12036–12040. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price M A, Tullius T D. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- 8.Koren R, Mildwan A S. Biochemistry. 1977;16:241–249. doi: 10.1021/bi00621a013. [DOI] [PubMed] [Google Scholar]
- 9.Jin D J, Gross C A. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 10.Severinov K, Soushko M, Goldfarb A, Nikiforov V. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 11.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S, Goldfarb A. J Biol Chem. 1995;270:23926– 23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 12.Weilbaecher R, Hebron C, Feng G, Landick R. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 13.Landick R, Stewart J D, Lee N. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 14.Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Kashlev M, Borukhov S, Goldfarb A. Proc Natl Acad Sci USA. 1991;88:6018–6022. doi: 10.1073/pnas.88.14.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kashlev M, Lee J, Zalenskaya K, Nikiforov V, Goldfarb A. Science. 1990;248:1006–1009. doi: 10.1126/science.1693014. [DOI] [PubMed] [Google Scholar]
- 17.Sagitov V, Nikiforov V, Goldfarb A. J Biol Chem. 1993;268:2195–2202. [PubMed] [Google Scholar]
- 18.Treich I, Carles C, Sentenac A, Riva M. Nucleic Acids Res. 1992;20:4721–4725. doi: 10.1093/nar/20.18.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grachev M, Lukhtanov E, Mustaev A, Zaychikov E, Abdukayumov M, Rabinov I, Richter V, Skoblov Yu, Chistyakov P. Eur J Biochem. 1989;180:577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
- 20.Mustaev A, Kashlev M, Lee J, Polyakov A, Lebedev A, Zalenskaya K, Grachev M, Goldfarb A, Nikiforov V. J Biol Chem. 1991;266:23927–23931. [PubMed] [Google Scholar]
- 21.Severinov K, Mustaev A, Severinova E, Kozlov M, Darst S, Goldfarb A. J Biol Chem. 1995;270:29428–29432. doi: 10.1074/jbc.270.49.29428. [DOI] [PubMed] [Google Scholar]
- 22.Borukhov S, Lee J, Goldfarb A. J Biol Chem. 1991;266:23932–23935. [PubMed] [Google Scholar]
- 23.Markovtsov V, Mustaev A, Goldfarb A. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severinov K, Mustaev A, Severinova E, Bass I, Kashlev M, Landick R, Goldfarb A. Proc Natl Acad Sci USA. 1995;92:4591–4595. doi: 10.1073/pnas.92.10.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst S, Goldfarb A. J Biol Chem. 1996;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 26.Mustaev A, Kashlev M, Zaychikov E, Grachev M, Goldfarb A. J Biol Chem. 1993;268:19185–19187. [PubMed] [Google Scholar]
- 27.Young X, Price C W. J Biol Chem. 1995;270:23930–23933. doi: 10.1074/jbc.270.41.23930. [DOI] [PubMed] [Google Scholar]