Summary
Specialized subsets of T lymphocytes can distinguish the carbohydrate portions of microbial and self-glycolipids when they are presented by proteins in the CD1 family of antigen presenting molecules. Recent immunochemical and structural analyses indicate that the chemical composition of the presented carbohydrate, together with its precise orientation above the CD1 binding groove, determines if a particular T cell is activated. More recently, however, it has been shown that the lipid backbone of the glycolipid, buried inside the CD1 protein, also can have an impact on T cell activation. While glycolipid recognition is a relatively new category of T cell specificity, the powerful combination of microbial antigen discovery and structural biochemistry has provided great insight into the mechanism of carbohydrate recognition.
Introduction
The highly diverse antigen receptors of B and T lymphocytes are formed by somatic recombination of the variable (V) and joining (J) DNA segments encoding them. Although a lymphocyte expresses only a single antigen receptor, diversity in the population confers upon lymphocytes the capacity to recognize an almost infinite number of different antigens. While the antigen receptors expressed by B lymphocytes can recognize all types of antigens, most T cells recognize peptides that are bound to the groove of a class I or class II cell surface protein encoded by genes in the major histocompatibility complex (MHC). T cell antigen recognition is therefore mostly confined to antigenic peptide fragments that are bound to cell surface proteins.
Cesar Milstein and collaborators originally defined a third family of antigen presenting molecules, the CD1 molecules [1]. CD1 proteins have a groove similar to the MHC-encoded antigen presenting molecules, but this groove is highly hydrophobic [2]. Therefore, CD1 presents hydrophobic antigens, mostly glycolipids, with the hydrophilic carbohydrate portion protruding from the CD1 groove and available for recognition by the T cell antigen receptor (TCR). In this review, we consider recent advances in identifying glycolipid antigens presented by CD1 molecules, in understanding how these bind to CD1 molecules, and finally, how the glycolipid-CD1 complexes might be recognized by the TCR.
CD1 genetics and structure
CD1 molecules are cell surface glycoproteins expressed mainly by white blood cells, including B lymphocytes, macrophages and dendritic cells (DC). They consist of two chains: β2 microglobulin (β2m), also found in MHC class I molecules, and a heavy chain containing three extracellular domains (α1-α3). The α1-α2 super domain forms the antigen-binding groove and it consists of two α-helices (α1 and α2) that sit atop a 6-stranded anti-parallel beta-sheet platform. The membrane proximal α3 domain binds β2m (Figure 1A, lower panel). The number of CD1 genes varies with the species. Humans have five CD1 isotypes (CD1a–e), which can further be divided in three groups (CD1a–c, CD1d and CD1e), while mice and rats have only CD1d [3,4].
Fig. 1.
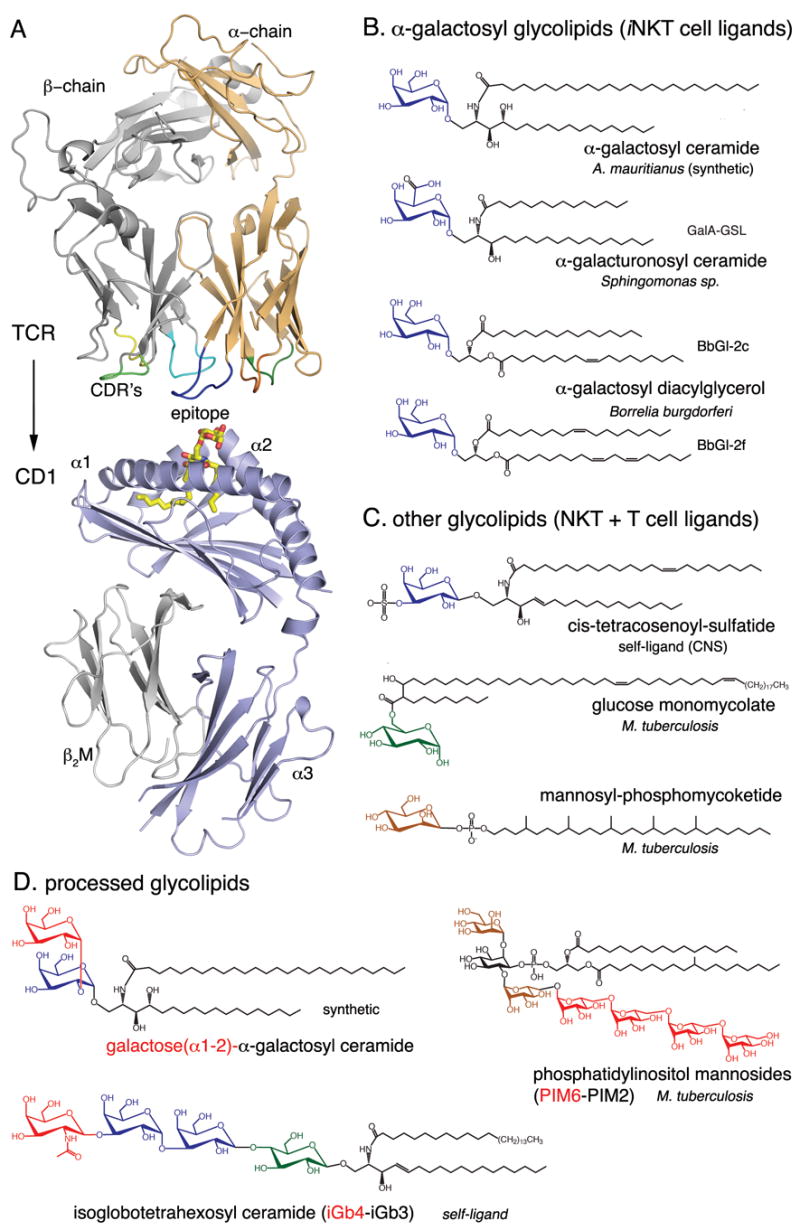
Structures of CD1d, the invariant TCR and glycolipid ligands. (A) Schematic ribbon diagram representation of the protein backbone of a human Vα24-Vβ11 iNKT cell TCR (top) and mouse CD1d (bottom). The α-galactosylceramide ligand (α-GalCer, yellow) binds with the lipid backbone inserted into the binding groove, while the carbohydrate epitope is exposed at the CD1d surface for recognition by the complementarity determining regions (CDR’s) of the TCR. (B–D) Chemical structures of some of the glycolipid ligands presented by CD1 molecules. The origin of the ligand is indicated, as well as the CD1 presenting molecule. (B) iNKT cell ligands presented by CD1d containing galactose or modified galactose bound with an α-glycosidic linkage to either a ceramide or diacylglycerolipid backbone. (C) Epitopes of other self and microbial antigens including. (D) Some carbohydrate epitopes that require lysosomal processing in order to be recognized by T cells. Carbohydrates in red are removed during processing. Otherwise, galactose is blue, glucose is green and mannose is brown. Figures were prepared with PyMol using the PDB coordinates 1Z5L and 2EYR.
Glycolipid reactive T cells
It is known from structural and other studies that the TCR of peptide reactive T cells typically makes contacts with the exposed side chains of the bound peptide antigen together with α-helical amino acids of the class I or class II molecule. It is likely that the glycolipid-reactive TCRs behave similarly, binding to CD1 together with the surface exposed, predominantly carbohydrate portion of the bound glycolipids (Figure 1A, upper panel).
Natural killer T (NKT) cells are the most widely studied population of glycolipid reactive lymphocytes. They were named because they express a TCR consisting of α and β chains, similar to most other T lymphocytes, as well as receptors typically found on NK cells, an innate immune cell type. Many NKT cells are specific for CD1d when it presents several types of glycolipids (Figure 1), and they express an invariant α chain rearrangement, Vα24 in humans and Vα14 in mice, that pairs with a limited number of β chains, primarily Vβ11 in humans and Vβ8.2 Vβ7 and Vβ2 in mice. Because of the invariant α chain, Vα24 or Vα14 as noted above, these cells are often termed invariant (i) NKT cells [5,6]. There are also CD1d reactive T cells that have more variable TCRs, however, and TCR variability is characteristic of human T cells reactive with the group I CD1 molecules.
CD1-presented glycolipid antigens
CD1 presented glycolipid antigens can be grouped into different classes including, but not limited to, diacylglycerolipids, sphingolipids, mycolates and phosphomycoketides (Figure 1 B–D) [2,7–9]. Microbial antigens from pathogenic mycobacteria, such as glucose monomycolates (GMM), mannosyl phosphomycoketides (Figure 1C) and phosphatidylinositol mannosides (PIM2 and PIM6, Fig. 1D), are known as potent ligands for human T cells when presented by group I CD1 molecules [10].
Most of the biological data on iNKT cell activation has been obtained using a glycosphingolipid called α-galactosyl ceramide (α-GalCer) obtained from the marine sponge Agelas mauritianus, (Figure 1B). This compound was discovered in a screen for natural anticancer activity, and was modified slightly by medicinal chemistry. Later it was shown that the α-GalCer anti-tumor activity resulted from the CD1d-dependent activation of iNKT cells [11]. Very recently, structurally related microbial α-glycuronosyl ceramides from Sphingomonas sp. and α-galactosyl diacylglycerols (BbGl-2) from Borrelia burgdorferi (Figure 1B) have been identified as both mouse and human iNKT cell antigens, emphasizing a role for these lymphocytes in host defense [12–15]. The striking feature of all of these iNKT cell agonists is their α-linked hexose sugar, while most mammalian glycolipids have β–linked carbohydrates attached to the lipid backbone. An interesting exception to this requirement for α linkage of the sugar to the lipid is the mammalian self-glycosphingolipid iGb3 (Fig. 1D), which has been shown to activate both human and mouse iNKT cells [18]. iGb3 contains a trisaccharide, and although the lipid proximal glucose is β linked, there is a terminal galactose that is β linked. This terminal α-linked galactose is required for antigenic activity, suggesting a surprising cross reactivity of the mono- and trisaccharide glycosphingolipid antigens. It has been proposed that recognition of iGb3 in the thymus and periphery is required for the normal development and function of mouse iNKT cells [16,17], but this hypothesis has been seriously challenged by several studies [18,19].
CD1 sorting and glycolipid processing
The primary function of CD1 is to survey the glycolipid content of antigen-presenting cells, such as B cells, macrophages and DC. The lipids can be loaded into the CD1 binding groove only if they meet the structural requirements for binding to the particular CD1 isotype. Lipid transfer proteins can assist in CD1 loading, including the lysosomal proteins saposin A–D, ganglioside GM2 activator, and Niemann-Pick disease protein type C2 (NPC2), and the ER-resident protein microsomal triglyceride transfer protein (MTTP) [20–28]. The exact mechanism of how these transfer proteins aid in glycolipid loading of CD1 molecules remains to be elucidated. Some glycolipids have more complex carbohydrate moieties (Figure 1D) that require carbohydrate processing, or degradation by lysosomal enzymes, to generate smaller, lipid-linked sugars that can be recognized by TCRs when bound to CD1. CD1e, whose function remained elusive, recently has been found to play a role in lipid loading and carbohydrate processing. The proposed model suggests that CD1e binds glycolipids, such as phosphatidylinositol hexamannosides (PIM6), and that CD1e activates lysosomal α-mannosidases that further process hexamannosylated PIM6 to di-mannosylated PIM2, which can then be presented by CD1b and recognized by antigen-specific TCRs [29].
CD1-glycolipid complexes
Over the last two years, various crystal structures have been determined for several CD1 molecules bound to either α-GalCer [30,31], α-galacturonosylceramide (GalA-Gsl) [32], phosphatidylcholine (PC) [33,34], sulfatide [35] and phosphatidylinositol di-mannoside (PIM2) [36] (Figure 2). In contrast to the group I CD1-glycolipid structures, which include CD1a with bound sulfatide [37] (Figure 2A) and CD1b with bound GMM [38] (Figure 2B), the CD1d glycolipid structures revealed an extensive hydrogen-bonding network that is formed with the polar moieties of the antigen.
Fig. 2.
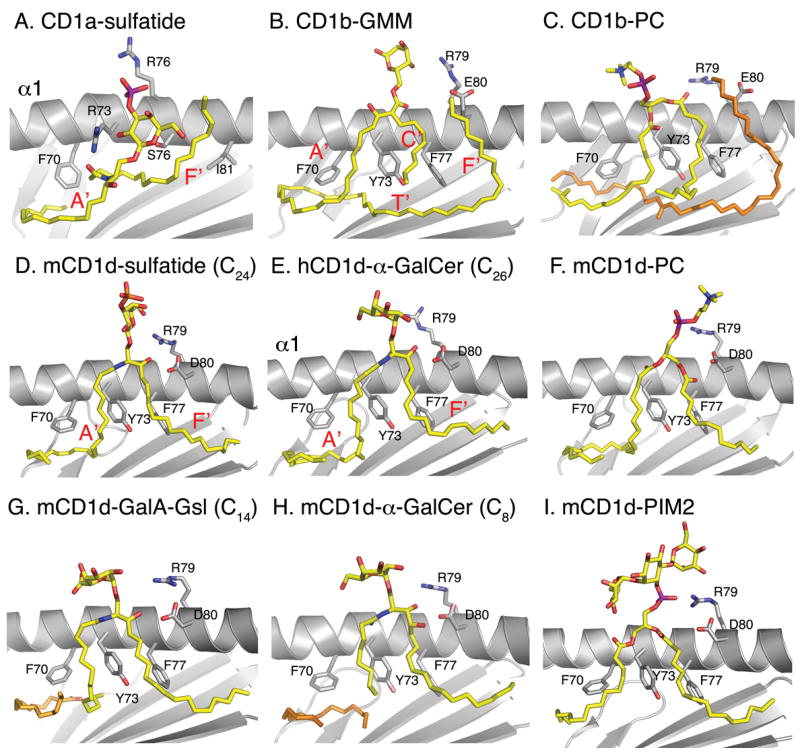
Overview of ligand binding to CD1 molecules. A side-view of recent crystal structures of CD1d (grey) in complex with different ligands (yellow) is depicted, compared with two earlier structures of human CD1a and CD1b. The α2-helix is removed for clarity, and the side chains of some key amino acids are indicated. The locations of the different CD1 binding pockets (A′, C′, F′, T′) are shown in red letters. Spacer lipids that fill the CD1 grooves (2C, 2H) are shown in orange. CD1a and CD1b ligands are mainly bound such that the hydrophobic lipid backbone is stabilized by non-polar van der Waals interactions with non-polar amino acids that line the interior or the CD1 groove, while the CD1d presented lipids (D, E and G–I) are bound by a well defined hydrogen-bond network with indicated charged CD1d residues Arg79, Asp80, Asp153 and Thr156 (see Figure 4 for detail).
A T cell view of CD1-glycolipid complexes
Different glycolipids vary in the presented carbohydrate epitope and the mode of presentation (Figure 3). The sulfogalactosyl moiety of sulfatide presented by CD1a is mostly at the opening of the CD1 binding groove, barely above the α helices. When mouse CD1d presents sulfatide, however, the sulfogalactosyl moiety sits above and across the binding groove and is therefore highly exposed compared to sulfatide bound to CD1a (see Figs 2A and 2D and 3A and 3D). This indicates that the antigenic structure presented to the T cell is not determined solely by the chemical composition of the ligand, but that the CD1 isotype also plays a role.
Fig. 3.
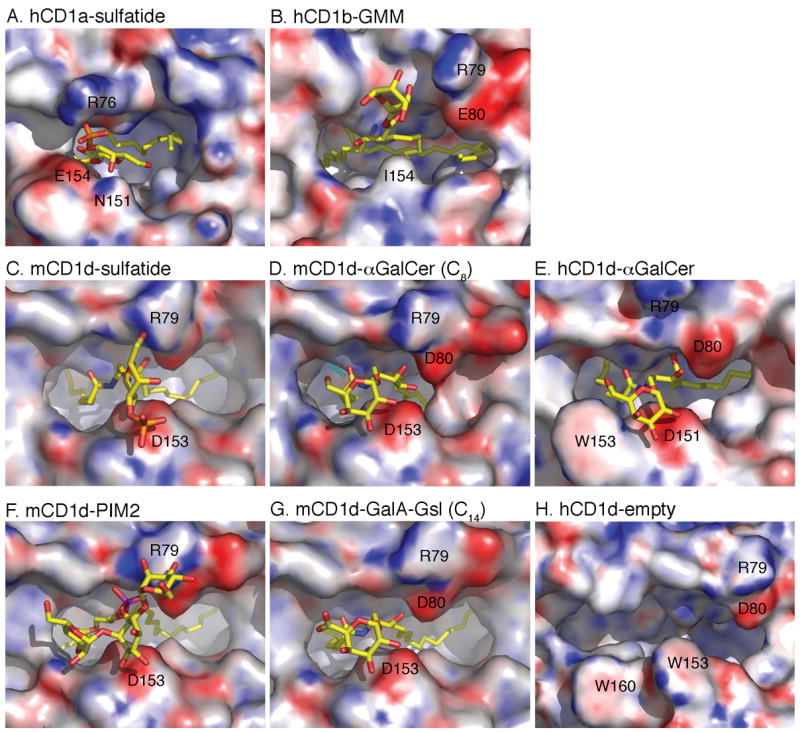
A TCR view of various CD1-glycolipid complexes. Looking down on the top of the molecule, the CD1 surface is shown with the calculated electrostatic surface potentials at pH 7.0 (blue is electropositive and red is electronegative, −15/+15 kT/e). The yellow glycolipids protrude from the groove towards the incoming TCR. Note how the lipid backbone is almost exclusively buried inside the CD1 groove, while the carbohydrate serves as the major T cell epitope. Binding of the highly stimulatory iNKT cell ligand α-GalCer to mouse CD1d (D) leads to an induced fit, which results in the formation of a roof next to D80 and D153. This partial closure is less pronounced in the comparable human structure (E) but it is quite different from the wide shaped binding groove that was observed in vitro during refolding experiments (H).
All CD1d presented α-linked glycosphingolipids are presented in a similar fashion, but subtle differences exist that are discussed in the next section. The induced fit upon binding of α-GalCer to CD1d is clearly visible in the mouse CD1d surface (Figure 3D), as are the more dramatic conformational changes between empty and α-GalCer loaded human CD1d molecules. PIM2 has the most complex carbohydrate epitope of all crystallized CD1 ligands, and it sits above the N-terminal half of the CD1d binding groove, while each sugar forms polar contacts with CD1 residues (Figure 3F). Although PIM2 has been considered a CD1b ligand, it is a basic building block of PIM4, which has been suggested as an antigen for a minority of mouse iNKT cells [39].
T cell recognition of α-linked glycosphingolipids
Earlier mutational studies [40] have demonstrated a crucial role for several CD1d residues for iNKT cell activation. All of these residues have now been shown through structural analysis to either participate in the hydrogen-bond network that stabilizes the antigenic head group of the glycolipid for proper TCR engagement (Arg79, Asp80, Asp153 and Thr156, see Fig. 4), and/or they are predicted to directly interact with the TCR (Glu83, Arg79) to facilitate CD1d-lipid antigen-TCR ternary complex formation [40]. Asp80 in the α1-helix interacts with the 3-OH group of the sphingosine base, whereas Asn153 in mouse CD1d (151 in human CD1d) stabilizes the head groups through interaction with either the 2′- and/or the 3′-OH groups of the sugars. Thr156 mouse CD1d (Thr154 in human CD1D) interacts with the oxygen of the O-glycosidic linkage and with the sphingosine backbone nitrogen. Additional residues, such Arg79 in mouse CD1d, can provide additional specificity for the ligand, but are also in a suitable location for interacting with the incoming TCR
Fig. 4.
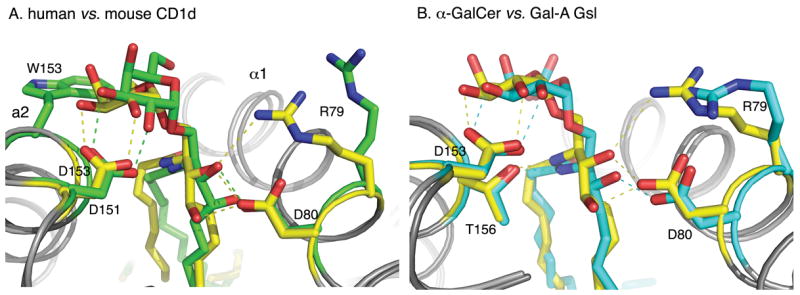
CD1d hydrogen-bonding network. The CD1d ligands (yellow, green and cyan) are stabilized by hydrogen-bonds (dashed lines) between the α1 and α2-helices of either human CD1d (A) or mouse CD1d (B). (A) Comparison of α-GalCer presentation by mouse (yellow) and human (green) CD1d. The ligands are in the same color as the CD1d molecules they are bound to, in order to emphasize the overall conformational change in the ligand-CD1d complexes. The most dramatic difference is W153 in human (G155 in mouse), which tilts and pushes the galactose towards the center of the CD1d binding grove. (B) Comparison of α-GalCer (yellow) and GalA-Gsl (cyan) binding to mouse CD1d. GalA-Gsl is inserted deeper into the F′ pocket and, as a result, the galacturonic headgroup is laterally shifted 1 Å towards the center of the binding groove.
Interestingly, mouse and human CD1d are very similar in structure and in their binding of α-GalCer, and in fact there is interspecies cross reactivity between mouse and human iNKT cells [41]. The only major difference occurs around Trp153 in the human ortholog, which raises and tilts the galactose head group slightly, whereas the corresponding mouse residue is the much smaller glycine, which does not influence ligand binding (Figure 4 A).
Both α-GalCer and the Sphingomonas glycolipid GalA-Gsl activate iNKT cells, but α-GalCer is a much more potent antigen. Most of the polar interactions between the two ligands and mouse CD1d residues are conserved; however, slight structural differences, such as the lack of the sphingosine 4-OH group in GalA-Gsl, affect the fine positioning of the ligand in the binding groove. Compared to α-GalCer, GalA-Gsl lacks the hydrogen bond with Asp80 and, as a result, the sphingosine chain is inserted slightly deeper into the F′ pocket (Figure 4B). This altered interaction results in an overall tilt of GalA-Gsl in the binding groove, which leads to lateral shift of the galacturonosyl head group by about 1 Å along the CD1d surface. This tilt in turn could lead to a different interaction with the TCR and, hence, could explain the weaker T cell stimulation. α-GalCer also induces slight, but potentially important structural changes in the α1-helix, which cause a more intimate association with the ligand (Figure 3D). It was proposed that the two hydrogen bonds between the 3-OH and 4-OH of the sphingosine base of the ceramide lipid of α-GalCer and CD1d amino acid Asp80 are responsible for pulling the α1-helix toward the ligand. The induced structural changes result in the formation of a roof above the F′ pocket, which increases the T cell recognition surface and, could provide additional avidity for the TCR.
Structure of glycolipid reactive TCRs
Immunoglobulin (Ig) and TCR V regions share a common structure, as they are composed of a series of anti-parallel β strands. The loops between several of these strands make up the complementarity determining residues (CDRs) that are responsible for antigen bindings. There are three CDRs in the α chain, and three in the α chain. The crystal structures of human Vα24 iNKT cell TCRs, paired with Vβ11 [42,43] and two other α-GalCer reactive, non-Vα24+ TCRs [43], have been reported. Although ternary CD1d-lipid-TCR crystal structures are not available yet, docking models have been proposed based on the MHC-peptide-TCR structures, which show a canonical diagonal TCR orientation onto the MHC surface. In the models for CD1d-glycolipid binding, the CDR3α, CDR3β and CDR1β contact the ligand, while CDR2β binds to the α1-helix [43]. One of the two crystallized Vα24 TCRs displayed a positively charged, pre-formed binding pocket composed of residues from CDR1α and CDR3α from the α-chain and CDR1β and CDR3β from the β-chain, which could accommodate and interact with the galactose head group of α-GalCer [42] and perhaps iGb3 as well. In addition, the positive charge of the TCR binding pocket would be well suited to neutralize the additional negative charge of the galacturonosyl head group. Although these structures give important structural insights into the NKT cell recognition of CD1d-presented glycolipids, the current lack of a CD1d-α-GalCer -TCR ternary complex makes it difficult to formulate detailed predictions about glycolipid recognition, especially to explain the observed differences in biological response to α-GalCer and GalA-Gsl.
The surprising role of the lipid backbone
It has become apparent that the positioning of the carbohydrate portion of the glycolipid, in the context of the presenting CD1 isotype, determines the ability of the antigen to activate T cells. α-GalCer is the most potent iNKT cell antigen, and therefore the position of the galactose above the mouse CD1d binding groove can be considered optimal, while any deviation from that position results in a reduced antigenic potency, for example 10–100 fold decreased for GalA-Gsl. However, there is now a growing body of evidence that slight alterations of the lipid backbone, such as changes in lipid chain length and saturation, have a dramatic impact on T cell recognition. The Borrelia α-galactosyl diacylglycerolipids BbGl-2c and BbGl-2f differ only slightly in lipid chain length and unsaturation (Figure 1B), however, while BbGl-2c stimulates mouse iNKT cells when presented by mouse CD1d, BbGl-2f does not [15]. The scenario is reversed for human iNKT cells, which prefer BbGl-2f and do not respond to BbGL-2c. Although none of these subtle changes have yet been captured at the molecular level through crystal structures, these minor changes may affect the positioning of the carbohydrate epitope and/or the conformation of CD1d, which could dramatically affect the quality of the T cell signal. A recent analysis of the biochemical basis for the response of human iNKT cells to variants of α-GalCer has been particularly illuminating in showing the importance of lipid structure [44]. Reducing the aliphatic chain length, of either the acyl chain, bound to the A′ pocket or the sphingosine bound to the F′ pocket (Figure 2), could affect the stability of lipid CD1d binding, as could the addition of two unsaturated bonds to the acyl chain. However, only the sphingosine changes could greatly alter the TCR affinity, suggesting that a full length, 18 carbon phytosphingosine, such as that found in α-GalCer, is required for optimal TCR affinity. The authors suggest that the sphingosine is needed to induce the closing of the CD1d helices over the F′ pocket observed with α-GalCer, but not in either empty human CD1d [31] or when mouse CD1d binds the Sphingomonas glycolipid GalA-GSL [32].
Conclusions
Although glycolipid recognition is a relatively new paradigm in studies of T cell specificity, progress has been rapid in uncovering the mechanism underlying the fine specificity of the recognition of antigens containing carbohydrates. The recent work shows that the TCR may be capable of reading not only the carbohydrate structure, but also subtle differences in the lipid antigen structure by sensing conformational changes in CD1. As more microbial glycolipid responses and antigens are studied, it will be interesting to determine if microbes evade CD1-mediated antigen recognition by altering the fatty acids they incorporate into glycolipids.
Intriguing questions regarding T cell recognition still remain unanswered in the absence of a tri-molecular structure that includes the TCR bound to a CD1-glycolipid complex. We do not know how the TCR contacts CD1. Of particular interest are the contact points for the highly conserved and selected invariant α chain. Furthermore, how is it possible that a TCR with a conserved α chain can on one hand recognize related antigens, such as the α-galactosyl containing glycolipids, while on the other hand, the more complex glycolipid iGb3 is recognized as well? Therefore, the structural determination of the different CD1-glycolipid complexes bound to their respective TCRs is one of the next major steps in elucidating the biology of glycolipid recognition by T cells.
Acknowledgments
Supported by NIH grants AI45053 and AI71922. This is publication number 971 from the La Jolla Institute for Allergy & Immunology.
Abbreviations
- α-GalCer
α-galactosyl ceramide
- GalA-Gsl
α-galacturonosylceramide
- β2m
β2 microglobulin
- CDR
complementarity determining residues
- DC
dendritic cell(s)
- GMM
glucose monomycolates
- i
invariant
- Ig
immunoglobulin
- J
joining segment of rearranging antigen receptor genes
- MHC
major histocompatibility complex
- NKT
natural killer T
- PC
phosphatidylcholine
- PIM
phosphatidylinositol mannosides
- TCR
T cell antigen receptor
- V
variable
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note added in proof: After the submission of this article, the tri-molecular structure of a human Vα24i NKT cell TCR bound to a human CD1d complex with α-GalCer was published (Borg NA, Wun KS, Kjer-Nielsen L, Wilce MCJ, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J: CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 2007, 448:44-49). The authors show that the invariant TCR has an unusual orientation to the CD1d-groove, focused on one end of CD1d and parallel to the long axis of the groove, as opposed to the diagonal orientation to MHC class I and class II typical of peptide reactive TCRs. Moreover, the main TCR contacts with the CD1d-glycolipid complex are mediated by the invariant α chain CDR1 and CDR3 regions, with TCR β chain contacts limited to CDR2 of the β chain contacting the C-terminal end of the CD1d α1 helix.
References
- 1.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 2.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 3.Dascher CC, Hiromatsu K, Naylor JW, Brauer PP, Brown KA, Storey JR, Behar SM, Kawasaki ES, Porcelli SA, Brenner MB, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- 4.Van Rhijn I, Koets AP, Im JS, Piebes D, Reddington F, Besra GS, Porcelli SA, van Eden W, Rutten VP. The Bovine CD1 Family Contains Group 1 CD1 Proteins, but No Functional CD1d. J Immunol. 2006;176:4888–4893. doi: 10.4049/jimmunol.176.8.4888. [DOI] [PubMed] [Google Scholar]
- 5.Kronenberg M. Toward an Understanding of NKT Cell Biology: Progress and Paradoxes. Annu Rev Immunol. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 6.Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 7.De Libero G, Mori L. Recognition of lipid antigens by T cells. Nat Rev Immunol. 2005;5:485–496. doi: 10.1038/nri1631. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji M. Glycolipids and phospholipids as natural CD1d-binding NKT cell ligands. Cell Mol Life Sci. 2006;63:1889–1898. doi: 10.1007/s00018-006-6073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bricard G, Porcelli SA. Antigen presentation by CD1 molecules and the generation of lipid-specific T cell immunity. Cell Mol Life Sci. 2007 doi: 10.1007/s00018-007-7007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brigl M, Brenner MB. CD1: Antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi E, Motoki K, Uchida T, Fukushima H, Koezuka Y. KRN7000, a novel immunomodulator, and its antitumor activities. Oncol Res. 1995;7:529–534. [PubMed] [Google Scholar]
- 12.Sriram V, Du W, Gervay-Hague J, Brutkiewicz RR. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur J Immunol. 2005;35:1692–1701. doi: 10.1002/eji.200526157. ●●This paper shows that Sphingomonas bacteria, commonly found in the environment, contain glycosphingolipids with α linked sugars that are antigenic for iNKT cells, thereby demonstrating that iNKT cells have microbial reactivity. Also see references 13 and 14. [DOI] [PubMed] [Google Scholar]
- 13.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. ●● In addition to demonstrating iNKT cell glycosphingolipid reactivity, here it is shown that mice lacking iNKT cells have delayed Sphingomonas clearance. See also reference 14. [DOI] [PubMed] [Google Scholar]
- 14.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. ●●. [DOI] [PubMed] [Google Scholar]
- 15.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. ●● This paper shows that pathogenic Borrelia bacteria have antigens for iNKT cells. The antigens are glycosylated diacylglycerol lipids, indicating that compounds besides glycosphingolipids can activate these cells. It is also demonstrated that subtle alterations in lipid composition dramatically affect antigenic potency. [DOI] [PubMed] [Google Scholar]
- 16.Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 17.Zhou D. The immunological function of iGb3. Curr Protein Pept Sci. 2006;7:325–333. doi: 10.2174/138920306778018007. [DOI] [PubMed] [Google Scholar]
- 18.Speak AO, Salio M, Neville DC, Fontaine J, Priestman DA, Platt N, Heare T, Butters TD, Dwek RA, Trottein F, et al. Implications for invariant natural killer T cell ligands due to the restricted presence of isoglobotrihexosylceramide in mammals. Proc Natl Acad Sci U S A. 2007;104:5971–5976. doi: 10.1073/pnas.0607285104. ● The authors report that they could not detect the self-antigen iGb3 in mouse or human thymus or DC, although they could detect it in the same tissues from rats, and it also was found in the mouse dorsal root ganglion. Although the detection limit is an issue, these data suggest that iGb3 may not be an antigen required for iNKT cell differentiation and function. See also reference 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porubsky S, Speak AO, Luckow B, Cerundolo V, Platt FM, Grone HJ. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci U S A. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. ● Mice lacking iGb3 synthase were found to have a normal number of iNKT cells and a normal function of these cells, despite the expected absence of chemically detectable iGb3. In agreement with reference 18, these data suggest that iGb3 may not be an antigen required for iNKT cell differentiation and function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dougan SK, Salas A, Rava P, Agyemang A, Kaser A, Morrison J, Khurana A, Kronenberg M, Johnson C, Exley M, et al. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. ● The authors show that microsomal triglyceride transfer protein (MTP), an endoplasmic reticulum (ER) chaperone that loads lipids onto apolipoprotein B, also loads phospholipids to CD1d and is required for normal CD1d function in vivo. The data suggest that MTP acts as an ER chaperone for CD1d allowing it to be loaded with placeholder lipids that can be exchanged for more antigenic lipids in lysosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou D, Cantu C, 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 23.Schrantz N, Sagiv Y, Liu Y, Savage PB, Bendelac A, Teyton L. The Niemann-Pick type C2 protein loads isoglobotrihexosylceramide onto CD1d molecules and contributes to the thymic selection of NKT cells. J Exp Med. 2007;204:841–852. doi: 10.1084/jem.20061562. ● The authors show that the Niemann-Pick type C2 protein, a soluble constituent of lysosomes, is important both for the exchange of lipids bound to CD1d and for normal iNKT cell function in vivo. Therefore, proteins in addition to the saposins defined earlier could be important for normal CD1d lipid antigen binding in lysosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Major AS, Joyce S, Van Kaer L. Lipid metabolism, atherogenesis and CD1-restricted antigen presentation. Trends Mol Med. 2006;12:270–278. doi: 10.1016/j.molmed.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 25.van den Elzen P, Garg S, Leon L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. ● This paper reports that apolipoprotein E binds lipid antigens and delivers them by receptor-mediated uptake into endosomes for CD1-mediated antigen presentation. Furthermore, antigen-presenting cells such as DC can secrete apolipoprotein E as a mechanism to capture antigens for uptake. [DOI] [PubMed] [Google Scholar]
- 26.Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 27.Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, Chen D, Kaser A, Glickman J, Kuo T, Little A, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10:535–539. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- 28.Yuan W, Qi X, Tsang P, Kang SJ, Illaniorov PA, Besra GS, Gumperz J, Cresswell P. Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc Natl Acad Sci U S A. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. ●● The function of the human CD1e isoform was not known until here the authors provide data indicating that CD1e can bind to glycolipids in intracellular vesicles and moreover, that it can facilitate the lysosomal processing of the carbohydrate portion of glycolipids to generate smaller structures that can be recognized by T cells. [DOI] [PubMed] [Google Scholar]
- 30.Zajonc DM, Cantu C, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;8:810–818. doi: 10.1038/ni1224. ●● α-GalCer is an exceptionally potent agonist for the iNKT cell TCR. Here the authors report on the structure of mouse CD1d bound to a compound closely related to α-GalCer, but with a shorter acyl chain. They demonstrate a network of hydrogen bonds between CD1d and the more hydrophilic portions of the glycolipid that hold the galactose sugar in a rigid, horizontal orientation. They also show that a spacer lipid is required to fill the CD1d groove with lipid, as also seen in the structures in references 32 and 34. See reference 31 for a similar structure with human CD1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, et al. The crystal structure of human CD1d with and without α-galactosylceramide. Nat Immunol. 2005;8:819–826. doi: 10.1038/ni1225. ●● This paper provides a structure of human CD1d bound to α-GalCer and a structure of an apparently empty or unloaded human CD1d molecule. The structures illustrate how the potent agonist causes conformational change in human CD1d. [DOI] [PubMed] [Google Scholar]
- 32.Wu D, Zajonc DM, Fujio M, Sullivan BA, Kinjo Y, Kronenberg M, Wilson IA, Wong CH. Design of NKT-cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. PNAS. 2006;103:3972–3977. doi: 10.1073/pnas.0600285103. ● The first structure of a natural, bacterial antigen bound to mouse CD1d is reported in this paper. Comparison to the structure of mouse CD1d bound to an α-GalCer analog provides evidence for an induced fit of CD1d upon α-GalCer binding, which may explain the exceptional potency of this antigen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol. 2005;175:977–984. doi: 10.4049/jimmunol.175.2.977. ● Here the structure of mouse CD1d bound to phosphatidyl choline is reported. The phosphatidyl choline ligand was loaded into the CD1d molecule in the insect cells used to produce soluble CD1d, but it may be similar or identical to compounds loaded into the CD1d groove after CD1d biosynthesis in mammalian cells. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. Embo J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. ● This paper shows the complex of human CD1b bound to the endogenous antigen phosphatidyl choline. CD1b has the largest interior groove in the CD1 family, and spacer lipids fill the remainder of the pocket, suggesting that a stable CD1 confirmation requires filling all or most of the groove with lipid. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. ● This paper reports the structure of mouse CD1d bound to sulfatide, an autoantigen that stimulates CD1d reactive T cells with more diverse TCRs. Sulfatide is an autoantigen that potentially may be involved in regulating autoimmunity in the central nervous system, and the conformation of its β linked sugar clearly shows the difference between α and β linked sugars presented by CD1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA. Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. J Immunol. 2006;177:4577–4583. doi: 10.4049/jimmunol.177.7.4577. ● Here the structure of PIM2 complexed to mouse CD1d is reported. PIM2 is a mycobacterial antigen presented by human CD1b, and it may be related to antigens for iNKT cells. The structure of PIM2 bound to CD1d illustrates how glycolipids with more complex carbohydrates may be bound and presented to T cells. [DOI] [PubMed] [Google Scholar]
- 37.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 38.Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, Besra GS, Cerundolo V, Jones EY. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2004;172:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 39.Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S, Hurwitz R, Kursar M, Bonneville M, Kaufmann SH, et al. Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2004;101:10685–10690. doi: 10.1073/pnas.0403787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamada N, Iijima H, Kimura K, Harada M, Shimizu E, Motohashi S, Kawano T, Shinkai H, Nakayama T, Sakai T, et al. Crucial amino acid residues of mouse CD1d for glycolipid ligand presentation to Vα14 NKT cells. Int Immunol. 2001;13:853–861. doi: 10.1093/intimm/13.7.853. [DOI] [PubMed] [Google Scholar]
- 41.Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. Structural requirements for galactosylceramide recognition by CD1-restricted NK T cells. J Immunol. 1998;161:5124–5128. [PubMed] [Google Scholar]
- 42.Kjer-Nielsen L, Borg NA, Pellicci DG, Beddoe T, Kostenko L, Clements CS, Williamson NA, Smyth MJ, Besra GS, Reid HH, et al. A structural basis for selection and cross-species reactivity of the semi-invariant NKT cell receptor in CD1d/glycolipid recognition. J Exp Med. 2006;203:661–673. doi: 10.1084/jem.20051777. ●● The structures of two Vα24i NKT cell TCRs are reported. Along with those in reference 43, these are the first structures of glycolipid TCRs, and they provide insight into the basis for glycolipid antigen recognition. The authors propose that a central, positively charged cavity in the TCR may be involved in recognition of the exposed galactose head group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadola SD, Koch M, Marles-Wright J, Lissin NM, Shepherd D, Matulis G, Harlos K, Villiger PM, Stuart DI, Jakobsen BK, et al. Structure and binding kinetics of three different human CD1d-α-galactosylceramide-specific T cell receptors. J Exp Med. 2006;203:699–710. doi: 10.1084/jem.20052369. ●● The authors report on the structures of three TCRs that react with α-GalCer presented by CD1d, only one of which has the canonical Vα24i rearrangement. Comparison of the structures allowed the authors to generate a docking model for understanding how the α-GalCer reactive TCRs interact with the glycolipid when it is bound to CD1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA, Bossi G, Salio M, Denkberg G, Reddington F, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. ●● By using α-GalCer analogs with different alkyl chain length the authors could demonstrate that alterations in the lipid chain length, especially in the alkyl chain that is bound in the F′ pocket, can have a direct effect on TCR affinity. The authors propose a model in which the shorter lipid tails would lead to a partial collapse of the F′ pocket, thereby affecting the CD1 surface that interacts with the TCR and consequently changing the TCR-glycolipid-CD1 affinity. [DOI] [PMC free article] [PubMed] [Google Scholar]


