Abstract
The helix-loop-helix protein, E47, is essential for both B- and T-lineage development. Here we demonstrate that in vitro E47 and Notch signaling act in concert to promote T cell development from fetal hematopoieitic progenitors and to restrain development into the natural killer and myeloid cell lineages. The expression of an ensemble of genes associated with Notch signaling is activated by E47, and additionally, Notch signaling and E47 act in parallel pathways to induce a T lineage–specific program of gene expression. Enforced expression of the intracellular domain of Notch rescues the developmental arrest at the T cell commitment stage in E2A-deficient fetal thymocytes. Finally, we demonstrate that regulation of Hes1 expression by Notch signaling and E47 is strikingly similar to that observed during Drosophila melanogaster sensory development. Based on these observations, we propose that in developing fetal thymocytes E47 acts to induce the expression of an ensemble of genes involved in Notch signaling, and that subsequently E47 acts in parallel with Notch signaling to promote T-lineage maturation.
Hematopoietic stem cells (HSCs) continuously generate progenitor cells of distinct lineages, including T, B, NK, dendritic, myeloid, and erythroid cells. Long-term HSCs mature into short-term repopulating cells. The short-term repopulating cell compartment contains multipotent progenitor cells, of which a fraction has the ability to differentiate into common lymphoid progenitor cells (CLPs) (1–3). Many different bone marrow populations have been proposed to seed the adult thymus, including HSCs, multipotent progenitor cells, early lymphoid progenitors, and CLPs (1, 2, 4). Thymus-colonizing progenitors in the fetal ages, however, were shown to develop from different origins (5–7).
Once the progenitors enter the thymus and interact with Notch ligands expressed on the thymic epithelium, T cell development is initiated. The earliest T cell precursor cells, known as early T-lineage precursors, exhibit high expression of c-kit and lack the expression of the coreceptors CD4 and CD8 (8–10). Early T-lineage precursors are present in the CD44+CD25− (DN1) subset, which maintain the ability to differentiate to diverse cell types, including B, NK, and dendritic lineages, but have minimal myeloid potential. Progenitor cells become committed to the T cell lineage at the CD44+CD25+ (DN2) and the CD44−CD25+ (DN3) cell stages.
The E2A gene encodes for two isoforms, E12 and E47, generated through alternative splicing of exons encoding the DNA binding and dimerization domain (11). The E2A proteins are classified as class I HLH proteins also named E proteins (12). They include E12, E47, HEB and E2-2, and the Drosophila homologue, daughterless. daughterless is most closely related to E12 and E47. In B lineage cells, the predominant E-box–binding complex is comprised of E47 homodimers, whereas in thymocytes, E47 readily forms heterodimers with HEB (12, 13).
Thymocyte development is perturbed in adult E2A-deficient mice, albeit partially, at the DN1 stage (14). Enforced expression of Id3 in human T-lineage precursor cells and expression of a dominant-negative form of HEB arrest thymocyte development also at the DN stage (15–17). Several target genes, including pTα and RAG1/2, have been shown to be regulated by E2A and HEB in developing T-lineage cells (18, 19).
T-lineage commitment requires Notch1-mediated signaling (20–26). Four mammalian Notch receptors have been identified, designated as Notch1–4. Notch receptors are activated upon interacting with different ligands, named Delta-like 1 (DL1), Delta-like 3, Delta-like 4, Jagged1, and Jagged2. Binding of the ligand to the Notch receptor activates proteolytic cleavage, resulting in the release of the intracellular portion of the receptor (ICN) from the cellular membrane. ICN is transported to the nucleus, where it interacts with CSL (CBF1, RBP-Jκ, suppressor of Hairless Su(H)) to activate down-stream target gene expression. Among its targets are Hes1 and pTα (22, 24). In the thymus, T cell development in Notch1-deficient mice is blocked at the DN1 stage (27). In contrast, enforced expression of ICN promotes T cell development in the bone marrow, whereas B cell development is perturbed (28). Conditional inactivation of the CSL gene and ectopic expression of antagonists of Notch signaling, including Deltex1, Nrarp, and Lunatic Fringe, inhibit T cell maturation at a similar stage (29–32). Consistent with being a key target for Notch1-mediated signaling, thymocyte maturation in Hes1-deficient mice is blocked at an early stage (33). How Hes1 contributes to T-lineage developmental progression is still poorly understood.
The critical roles for E proteins and Notch signaling at the early stage have raised the question of how these two ancient regulatory modules interact to control thymocyte commitment. Initial studies have suggested that Notch signaling acts to modulate the transactivation activity of E47 (28, 34). Further studies have indicated that enforced Notch signaling together with mitogen-activated protein (MAP) kinase signaling readily promotes the degradation of E47, suggesting a linear relationship (35, 36). Collectively, these studies suggest that E proteins and Notch signaling act in a linear pathway.
Here we show that in vitro E47 and Notch signaling act in concert to promote thymocyte development from fetal hematopoieitic progenitors. Furthermore, E47 and Notch signaling act in synergy in fetal progenitor cells to restrain NK and myeloid development. E47 also induces the expression of a subset of genes involved in Notch-mediated signaling, including Notch3, Hes1, Hes5, Grg6, and, albeit modestly, Notch1, and E47 and Notch signaling act synergistically to induce the expression of pTα and CD7. An early block of T cell development in E2A-deficient fetal thymocytes is rescued, in part, by the overexpression of ICN. Based on these and previous observations, we suggest that in the fetal thymus E47 acts to induce the expression of an ensemble of genes involved in Notch signaling and additionally that it acts in concert with Notch signaling to induce a T lineage–specific program of gene expression.
RESULTS
Global analysis of gene expression profiles in E2A-deficient hematopoietic progenitor cells in response to E47 activity
Previously, we described E2A-deficient hematopoietic progenitor cells (HPCs) that exhibit T, NK, dendritic, myeloid, and erythroid lineage potential (37). To identify E47 target genes in hematopoietic progenitor cells, E2A-deficient HPCs were transduced with retrovirus carrying an E47–estrogen receptor (E47ER) fusion protein. Control cells were transduced with virus carrying the bHLH region but lacking the NH2-terminal transactivation domains. Both retroviral constructs also contained the coding sequence of hCD25 to allow rapid isolation of transduced cells (Fig. 1 A). 2 d after infection, the cells were cultured in the presence of S17 stromal cells and incubated with 4-hydroxytamoxifen (4-OHT) for a period of 6 h to activate the E47ER fusion protein. hCD25-positive cells were purified using magnetic beads, and RNA was isolated (Fig. 1 B). The resulting RNAs were converted into cDNA that in turn was used to synthesize biotinylated cRNA, which subsequently was hybridized to mouse oligonucleotide arrays. Fluorescence intensities for each probe were normalized to the signal from oligonucleotides representing a group of housekeeping genes. The normalized hybridization intensities from each of the samples were then compared with that of RNA derived from control infected E2A-deficient HPCs. We identified those oligonucleotides that had ratios of either >4.0 (representing candidates for positive regulation by E47) or <0.5 (representing genes potentially repressed by E47). We focused our analysis on a selected group of genes involved in cell survival, proliferation, and developmental progression. Among this focused group of genes, there were 53 whose expression was increased by a factor of at least fourfold (Fig. 1 C). Among these were, interestingly, a set of genes that are known to play essential roles in T cell development and cell growth, including MHC class II, IL-7Rα, syk, pTα, Hes1, LAT, Id2, Grg6, Bcl2A1b, CD7, and cyclin D3 (Fig. 1 C). 21 genes repressed by a factor of twofold or more include Bcl11b, RORα, and Gfi1b (Fig. 1 C).
Figure 1.
E47 target genes expressed in E2A-deficient hematopoietic progenitor cells. (A) E47ER and bHLHER retroviral constructs. The genes encoding E47 and human CD25 are shown. (B) Protocol used for enforced overexpression of E47 in E2A-deficient HPCs. (C) Target genes activated (left) or repressed (right) by the overexpression of E47. Values indicated on the horizontal axis show the fold induction or repression of E47 target transcript levels. Only changes of more than twofold for the repression and fourfold for the induction are indicated.
To validate the data obtained from the microarray analysis, mRNA was isolated from the transduced cells and analyzed by real-time PCR (Fig. 2 A). As predicted from the data obtained from the microarray analysis, pTα, IL-7Rα, and LAT transcript levels were significantly elevated in cells expressing E47 (Fig. 2 B). To determine whether E47 directly activates the expression of these target genes, cells were induced with 4-OHT in the presence of cycloheximide (CHX). Each of the targets, including pTα, IL-7Rα, and LAT, were activated in the presence of CHX, indicating that the induction of transcription by E47 was direct (Fig. 2 C). These data indicate that enforced E47 expression in hematopoietic progenitor cells both activates and inhibits the expression of genes associated with a T lineage–specific program of gene expression.
Figure 2.
E47 directly activates in E2A-deficient hematopoietic progenitor cells the expression of genes involved in Notch-mediated signaling. (A) Protocol used to examine regulation of target genes in E2A-deficient HPCs by E47. (B) pTα, IL-7Rα, and LAT mRNA levels in cells either transduced with E47ER or with bHLHER fusion protein. Transcript levels were determined using real-time PCR. Shown is the ratio of transcript levels of E47ER compared with the bHLHER normalized by the acidic ribosomal protein (ARP) mRNA expression. (C) E47 directly regulates pTα, LAT, and IL-7Rα in E2A-deficient HPCs. E2A-deficient cells were treated for a 6-h period in the presence of 4-OHT and CHX. Transcript levels were measured using real-time PCR. Indicated is the fold induction normalized to the ARP. (D) E47 activates the expression of a subset of genes involved in Notch-mediated signaling. Transcript levels were determined by real-time PCR. Shown is the ratio of transcript levels of E47ER compared with bHLHER normalized to the ARP. (E) E47 directly activates the expression of a subset of genes involved in Notch-mediated signaling. E2A-deficient cells were treated for a 6-h period in the presence of 4-OHT and CHX. Transcript levels were measured using real-time PCR. Indicated is the normalized fold induction. (F) E47 induces the expression of Notch-associated genes. E47 instead of E47ER fusion protein is transduced in E2A-deficient cells. Transcript levels were analyzed by real-time PCR after 24 h of infection. (G) E47 directly regulates the expression of Notch-associated genes independent of Notch-mediated signaling. E2A-deficient cells transduced with E47ER were induced with 4OHT and CHX for 6 h in the presence of γ-secretase inhibitor IX. Transcript levels were analyzed by real-time PCR and normalized to ARP mRNA levels. (H) Transcript levels of pTα, IL-7Rα, and LAT in E2A+/+, E2A+/−, and E2A−/− cells derived from fetal day 15 thymocytes. Transcript levels were analyzed by real-time PCR and normalized versus ARP. Black bars represent mRNA levels in E2A-deficient thymocytes. Gray bars represent mRNA levels in E2A+/− thymocytes. White bars represent mRNA levels in wild-type thymocytes. (G) Transcript levels of Hes1, Hes5, Notch1, and Notch3 in E2A+/+, E2A+/−, and E2A−/− thymocytes. RNA was isolated from day 15 fetal thymocytes. Transcript levels were analyzed by real-time PCR and normalized versus ARP. mRNA levels in E2A-deficient (black bars), E2A+/− (gray bars), and wild-type (white bars) thymocytes are shown.
E47 activates the expression of an ensemble of genes involved in Notch-mediated signaling
Unexpectedly, the bHLH protein, Hes1, was among the target genes induced by the expression of E47 (Fig. 1). To explore whether additional components of the Notch signaling module are regulated by E47, RNA was analyzed by real-time PCR for the expression of Hes1, Hes5, Notch1, Notch2, Notch3, NumbL, Grg3, Grg6, and Deltex1 (Fig. 2 D). Hes1 expression was readily induced by E47 activity, as measured by real-time PCR, consistent with the data obtained from the microarray analysis (Fig. 1). Deltex1, Hes5, and Notch3 were also activated by E47. Furthermore, Notch1 expression levels were modestly elevated by E47 (Fig. 2 D). Transcript levels of Groucho-related corepressors, Grg3 and Grg6, which act together with the Hes gene products to inhibit down-stream target gene expression, were also modulated by E47-mediated transactivation (Fig. 2 D). The expression of genes involved in Notch-mediated signaling was increased by the enforced expression of E47, even in the presence of CHX, indicating direct regulation by E47 (Fig. 2 E). These data indicate that enforced expression of E47 in E2A-deficient HPCs rapidly leads to the induction or elevation of components of the Notch signaling pathway.
To exclude the possibility that the E47ER fusion protein aberrantly activates target gene expression, we enforced the expression of E47 in the absence of an ER fusion component. We found that also in the absence of the estrogen domain E47 induces the expression of Hes1, Notch3, and Notch1 in E2A-deficient HPCs (Fig. 2 F).
Because E2A-deficient cells are grown on S17 cells, we considered the possibility that Jagged-mediated signaling would contribute to the induction of, for example, Hes1 expression by E47. To exclude this possibility, E2A-deficient cells transduced with E47ER were cultured in the presence of γ-secretase inhibitor IX. Hes-1, Notch1, and Notch3 mRNA levels were significantly induced by E47 in the presence of the γ-secretase inhibitor IX, indicating that the induction of the expression of these genes was not mediated by Notch signaling (Fig. 2 G).
To further define the involvement of E47 in the regulation of these genes, we examined the expression of these genes in fetal E2A+/+, E2A+/−, and E2A−/− thymocytes derived from day 15 embryos. RNA was isolated and analyzed by real-time PCR using the appropriate primers. pTα, IL-7Rα, and LAT transcripts were reduced in E2A-deficient thymocytes (Fig. 2 H). Hes1, Hes5, Notch1, and Notch3 transcript levels also were significantly lower in E2A-deficient thymi when compared with wild-type thymi (Fig. 2 I). E2A heterozygous thymocytes also showed a decrease in LAT, pTα, Hes1, and Hes5 mRNA levels as well, indicating that the regulation of these genes by E47 is dosage sensitive (Fig. 2, H and I). Collectively, these data indicate that in fetal thymocytes E2A proteins contribute to the regulation of a subset of genes involved in Notch-mediated signaling.
E2A directly regulates Hes1 gene expression
The observations described indicate that, surprisingly, Hes1 gene expression is activated by E47 in hematopoietic progenitor cells. Because Hes1 activation by E47 occurred in the presence of CHX, we conclude that E47 directly regulates Hes1 gene expression. The activation of Hes1 by E47 is interesting because it is reminiscent of the regulation of the Drosophila homologue of Hes1, enhancer of split, in Drosophila sensory organ development (38). Enhancer of split gene expression is regulated by daughterless, a basic helix-loop-helix (bHLH) protein strikingly similar to E47 (39). To identify the regulatory elements in the murine Hes1 locus that are responsive to E47 binding, we compared the DNA sequence of the human, murine, and chicken Hes1 loci. Four domains, named CR1-CR4, were identified that were highly conserved (Fig. 3 A). One of these conserved elements, CR3, contained two E47 high-affinity binding sites that are located 10 bp apart, in both the murine and human sequences, respectively (Fig. 3 A). To determine whether the CR3 element has the ability to activate gene expression, we inserted it upstream of a minimal promoter linked to the luciferase coding region. The reporter construct, as well as a construct lacking the CR3 element, was transfected into E2A-deficient HPCs in the presence of a vector carrying the E47 gene placed under control of the β-actin promoter. The reporter construct was activated by a factor of 2.5-fold compared with the reporter lacking the CR3 region (Fig. 3 C). To determine whether the increase in luciferase activity was dependent on the E47 binding sites, mutations were generated either in one of the sites (named E-box(1) and E-box(2)) or in both E-box sites (Fig. 3 B). Mutation of each of the binding sites substantially lowered luciferase activity (Fig. 3 C). These observations indicate that a highly conserved E47 responsive regulatory element is present in the Hes1 locus.
Figure 3.
E2A proteins bind to highly conserved E-box sites present in a highly conserved Hes1 regulatory element. (A) Alignment of human, mouse, rat, and chicken sequences surrounding the Hes1 locus. Vertical axis indicates over 50% homology compared with the human DNA sequence. Horizontal axis shows where homology is >50%. Four regions of substantial homology with chicken DNA sequence is shown, designated as CR1-CR4. Bottom panel shows the murine and human DNA sequences of the CR3 region. Identical nucleotides are indicated in gray. E-box sites are underlined. (B) DNA sequences of wild-type and mutant Hes-responsive elements used in transactivation studies are shown. (C) Normalized relative luciferase units of activity (RLU) using a dual luciferase reporter assay in E2A-deficient HPCs transfected with either vector alone or expressing E47. Constructs with the mutations in the E-box sites are indicated. (D) In vivo occupancy of the E-box sites in the Hes1 CR3 region in DN and DP thymocytes derived from wild-type and mice carrying an E2A–GFP fusion protein. Input DNA from each preparation is shown. An anti-GFP antibody was used to immunoprecipitate DNA fragments lysed DN and DP cells. DNA samples were serially diluted threefold and examined by PCR using primers flanking the CR3 region. (E) Analysis of E2A binding in vivo to the E-box sites present in the Hes1 CR3 region. Relative levels were determined using real-time PCR of the immunoprecipitated DNA fragments.
Hes1 is normally expressed at the double-negative (DN) cell stage but upon β-selection its levels decline significantly (33, 40, 41). To determine whether E47 binds to the CR3 region in DN and double-positive (DP) cells, we performed chromatin immunoprecipitation (ChIP) assay. Thymocytes from either wild-type mice or mice harboring a knock- in mutation in the E2A locus generating a E2A–GFP fusion proteins were sorted, and DN and DP populations were isolated, fixed with formaldehyde, and then disrupted by sonication (42). The cross-linked DNA was immunoprecipitated using a GFP-specific antibody. Immunoprecipitated DNA was amplified using Hes CR3 primers and examined by DNA gel electrophoresis. E47 binding to the Hes1 CR3 element was not significantly enriched in DP thymoctes derived from mice expressing the E2A–GFP fusion protein compared with wild-type thymocytes (Fig. 3 D). On the other hand, the CR3 element was substantially enriched in DN cells in E2AGFP mice (Fig. 3, D and E). Because Hes1 transcript levels are high in DN thymocytes but virtually absent in DP thymocytes, the binding activity of E47 to the Hes1 regulatory element correlates with Hes1 gene expression. These observations, in conjunction with the induction of Hes1 expression by E47 in the presence of CHX (Fig. 2 E), suggest that E47 directly regulates Hes1 gene expression by binding to a highly conserved regulatory element. These data imply that the molecular mechanism underlying enhancer of split gene expression has remained conserved in mammalian hematopoietic progenitor cells to regulate Hes1 transcription.
Notch-mediated signaling and E47 activate a common set of target genes
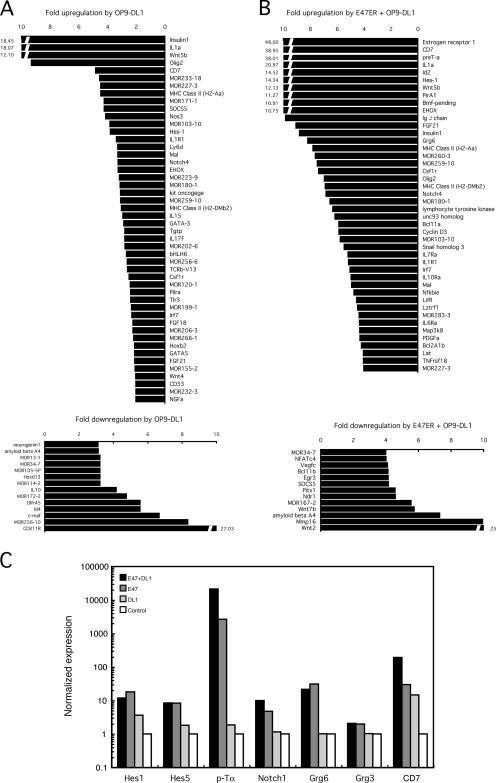
Previous data have established that Notch signaling activates the expression of Hes1 and Hes5 (21–24). Our data indicate that E47 directly activates Hes1, Hes5, and Grg6 expression. Collectively, the data raise the possibility that Notch signaling and E-protein activity act in parallel to induce target gene expression. To identify targets activated by Notch-mediated signaling alone and in conjunction with E47 expression, E2A-deficient HPCs were transduced with retrovirus carrying the E47 bHLH coding sequence. Transduced cells were cultured in the presence of stromal cells expressing the Notch ligand DL1 (OP9-DL1) or stromal cells expressing GFP (OP9-GFP) for a 6-h period (Fig. 4 A). The hCD25-positive cells were subsequently purified using magnetic beads, and RNA was isolated and analyzed for differences in down-stream target gene expression using microarrays. Interestingly, culturing E2A-deficient HPCs in the presence of the Notch ligand, DL1, showed elevated insulin and olfactory receptor transcript levels significantly within a 6-h period (Fig. 4 A). Additionally, GATA-3 and Hes1 mRNA levels were increased, consistent with them being targets for Notch-mediated signaling (24, 43). Notch signaling also activated the expression of CD7, MHC class II, EHOX, FGF21, and IRF7. We note that this set of target genes was also activated by the enforced expression of E47 alone.
Figure 4.
E47 and Notch signaling act in concert to regulate down-stream target gene expression in E2A-deficient hematopoietic progenitor cells. (A) Target genes activated or repressed in E2A-deficient cells upon exposure to the Notch ligand DL1 for a 6-h period. Values indicated on the horizontal axis show the fold induction (top) or repression (bottom). Only changes of more than threefold are shown. (B) Target genes activated or inhibited in E2A-deficient cells upon exposure to the Notch ligand DL1 and enforced expression of E47 for a 6-h period. Values indicated on the horizontal axis show the fold induction (top) or repression (bottom). Only changes of more than fourfold are shown. (C) E47 and Notch-mediated signaling act in concert to activate pTα gene expression in E2A-deficient progenitor cells. Transcript levels were analyzed by real-time PCR and normalized versus ARP. Black bars represent mRNA levels in E2A-deficient HPCs expressing E47 and exposed to Notch signaling. Dark gray bars represent mRNA levels in E2A-deficient HPCs expressing E47. Light gray bars represent mRNA levels in E2A-deficient HPCs exposed to Notch signaling. White bars represent mRNA levels in E2A-deficient HPCs cultured in the presence of OP9-GFP cells.
To identify target genes activated by both E47 and Notch signaling, E47ER-transduced cells were incubated for 6 h in the presence of 4-OHT and OP9-DL1. RNA was isolated from the transduced cells and examined by microarray analysis (Fig. 4 B). As expected, E2A-deficient cells exposed to both Notch signaling and E47 expression induced the expression of genes including Grg6, LAT, pTα, unc93 homologue, NFκBIɛ, PirA1, Bmf-pending, EHOX, Notch4, CD7, and Hes1, which also responded to either E47 and/or Notch-mediated signaling alone (Fig. 4 B). Notch signaling and E47 also acted to repress Mmp16, Wnt7b, Egr3, RORα, and Ndr1 expression (Fig. 1 and Fig. 4 B).
To examine whether Notch signaling and E47 act in synergy to induce the transcription of this set of genes, RNA was isolated from E2A-deficient HPCs carrying the E47ER fusion protein cultured in the absence or presence of Notch ligand, DL1. mRNA derived from these populations was analyzed by real-time PCR for the expression of Hes1, Hes5, pTα, Notch1, Grg3, and Grg6 (Fig. 4 C). As expected Hes1, Hes5, Notch1, and Grg6 transcription was activated by both E47 and Notch-mediated signaling, although the activation of gene expression was not synergistic (Fig. 4 C). However, E47 and Notch-mediated signaling acted in synergy to induce the expression of pTα and CD7 (Fig. 4 C). These data indicate that E47 activity and Notch signaling in E2A-deficient HPCs act in parallel pathways to induce the expression of a common set of target genes.
Enforced expression of Notch ICN rescues developmental arrest in fetal-derived E2A-deficient thymocytes
The data described in the previous paragraph suggest that E47 activates the expression of several components of the Notch signaling pathway and acts in parallel with Notch signaling to induce a program of T lineage–specific gene expression. In vivo, thymocyte development in adult E2A-deficient mice is partially blocked at the DN1 cell stage (14). However, a substantial fraction of thymocyte has the ability to develop into the mature T cell lineages (14). In contrast to adult thymocyte maturation, E2A-deficient fetal thymocytes inefficiently develop beyond the DN3 cell stage (unpublished data).
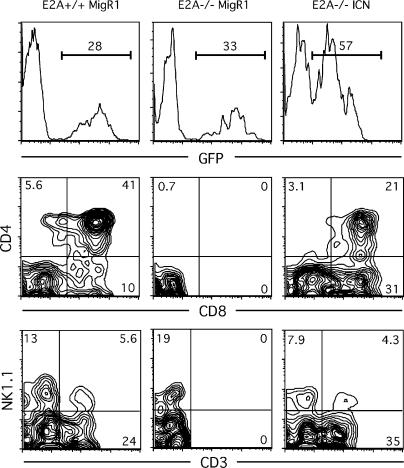
To determine whether constitutive active Notch signaling has the ability to overcome the impairment of T cell development in E2A-deficient fetal progenitors, we used a retrovirus vectors expressing ICN in fetal thymocytes. Hematopoietic progenitors derived from either E2A+/+ or E2A−/− fetal livers were infected with the retrovirus and cocultured with the deoxyguanosine-treated fetal thymic lobes. After 17 d of culture, the cells were harvested and analyzed by flow cytometry. As expected, E2A+/+ progenitors infected with control (MigR1) virus differentiated to CD4+CD8+ DP thymocytes as well as CD3−NK1.1+ NK cells (Fig. 5). On the other hand, E2A−/− progenitors infected with MigR1 virus gave rise to NK cells but failed to differentiate into CD3+ and DP cells (Fig. 5). These data are consistent with the phenotype of fetal E2A-deficient thymocytes that are severely perturbed in their ability to undergo T-lineage developmental progression. In contrast, enforced expression of ICN in fetal liver progenitor cells restored the ability of E2A-deficient progenitors to develop into CD3+ and DP cells. Collectively, these results indicate that the overexpresion of ICN can restore the impairment of T cell development of E2A-deficient progenitors.
Figure 5.
Rescue of T cell development in E2A-deficient fetal progenitors upon enforced expression of Notch-IC. Lin− fetal liver progenitors were transduced with control (MigR1) and Notch-IC (ICN) retroviral vectors and cultured with the deoxyguanosin-treated fetal thymic lobes (C57BL/6) for 17 d. Thymocytes were analyzed by flow cytometry for the expression of GFP, CD4, CD8, CD3, and NK1.1. The numbers in the histograms and quadrants represent the percentage of the cells. The data are representative of three independent experiments.
Notch signaling and E47 activity act together to promote T-lineage development at the expense of NK and myeloid cell maturation
The data shown in Fig. 4 indicate that Notch signaling and E47 have the ability to activate the expression of a common set of target genes. However, because the abundance of E47 expression from the retroviral construct is significantly higher than observed in wild-type thymocytes, the data do not provide unambiguous evidence that E47 and Notch signaling act in synergy to induce a T-lineage program of gene expression. To examine whether Notch signaling and E47 act in concert to promote thymocyte maturation, we cultured progenitors derived from E2A+/+, E2A+/−, and E2A−/− fetal liver in the presence of OP9-DL1 cells. Furthermore, to inhibit Notch-mediated signaling the cells were cultured in the presence of different concentrations of γ-secretase inhibitor IX.
E2A+/+ and E2A+/− progenitors efficiently developed into DN2 to DN3 stages in the absence of γ-secretase inhibitor (DMSO alone) after 7 d of culture (Fig. 5 A). In contrast, E2A−/− progenitors were severely impaired in their ability to differentiate into DN3 cells, raising the possibility that the ability of these cells to receive Notch signals is perturbed in the absence of E2A (Fig. 6 A). Consistent with previous observations, increasing concentrations of the γ-secretase inhibitor significantly perturbed thymocyte maturation, resulting at 10 μM concentration in a complete arrest at the DN1 stage (Fig. 6 A) (44). Interestingly, E2A+/− progenitors readily developed into the DN3 stage (54%) (Fig. 6 A). Similarly, a considerable proportion (23%) of E2A+/+ progenitors cultured in the presence of 3 μM γ-secretase inhibitor developed into the DN3 stage (Fig. 6 A). However, the majority of the E2A+/− cultured in the presence of 3 μM γ-secretase inhibitor was arrested at the DN1 stage and the fraction of DN3 cells (6.4%) was severely declined compared with wild-type thymocytes (23%) (Fig. 6 A). These results suggest that Notch signaling and E47 act in concert to promote developmental progression through the DN stage.
Figure 6.
E47 and Notch signaling act in concert to promote T cell development. Lin− progenitors (105 cells) derived from E14 E2A+/+, E2A+/−, and E2A−/− fetal livers were cultured on OP9-DL1 cells for 7 d with DMSO alone or in the presence of γ-secretase inhibitor IX at the indicated concentrations. (A) Flow cytometry analysis indicating the expression of CD25 and CD44 of fetal liver cells cultured on OP9-DL1 cells. The percentages of the number of cells present in the various compartments are indicated. (B) Flow cytometry analysis indicating CD19 and NK1.1 expression of cultured cells on OP9-DL1 cells. The percentages of the number of cells present in the quadrants are shown. (C) Total number of NK cells generated from 105 Lin− cells of E2A+/+, E2A+/−, and E2A−/− fetal livers cultured for 7 d on OP9-DL1 cells at the indicated concentration of γ-secretase inhibitor. The data are representative of three independent experiments.
To further define the effect of Notch signaling and E47 activity in lymphoid development, we also examined the generation of NK cells. In the E2A+/+ cells, a small number of NK cells were found in the absence of the Notch inhibitor (Fig. 6 B). However, the proportion of NK cells was significantly increased when cultured in the presence of γ-secretase inhibitors as previously demonstrated (Fig. 6 B) (44). Interestingly, E2A+/− and E2A−/− progenitors gave rise to substantial proportions of NK cells even in the absence of Notch inhibitor (Fig. 6 B). Moreover, the proportion and the absolute number of the NK cells were synergistically increased in the presence of Notch inhibitor in E2A+/− and E2A−/− progenitors (Figs. 6, B and C). These results suggest that Notch signaling and E47 act in concert to initiate T-lineage specification and to restrain fetal hematopoietic progenitors to adopt a NK cell fate.
To examine the effect of myeloid lineage development by the combined absence of E2A and Notch signaling, we analyzed the generation of Mac1+ cells (Fig. 7). A substantial fraction of E2A-deficient fetal progenitors developed into myeloid cells (Fig. 7 A). Interestingly, E2A+/− fetal hematopoietic progenitor cells showed an increased ability to differentiate into myeloid cells (Fig. 7 A). Strikingly, culturing E2A+/− progenitors in the presence of γ-secretase inhibitor IX synergistically increased the number of Mac1+ cells (Fig. 7 B). We note that the number of Mac1+ cells was significantly higher in cells derived from fetal hematopoietic E2A+/− compared with E2A−/− progenitors, suggesting that the E2A proteins play an essential role in myeloid lineage commitment and/or progression (Fig. 7 B).
Figure 7.
E47 and Notch signaling act in concert to induce T cell commitment at the expense of myeloid lineage development. Lin− progenitors (105 cells) derived from E14 E2A+/+, E2A+/−, and E2A−/− fetal livers were cultured on OP9-DL1 stromal cells for 7 d with DMSO alone or in the presence of γ-secretase inhibitor IX at the indicated concentrations. (A) Flow cytometry analysis indicating the expression of Mac1 and CD19. The percentages of the number of cells present in the various compartments are indicated. (B) Total number of Mac1+ cells generated from 105 Lin− cells of E2A+/+, E2A+/−, and E2A−/− fetal livers cultured for 7 d on OP9-DL1 cells at the indicated concentration of γ-secretase inhibitor.
Collectively these data suggest, in conjunction with the data described in the previous section, that in fetal thymocyte development E47 acts upstream of an ensemble of genes involved in Notch signaling. In the absence of E47, fetal thymocytes do not have the ability to receive and/or integrate Notch-mediated signaling and fail to differentiate into T-lineage cells but allow the development of NK and myeloid cells. During subsequent stages of fetal thymocyte maturation, E47 acts in parallel with Notch-mediated signaling to promote T-lineage developmental progression.
DISCUSSION
Previous data have indicated that the initial stages of B- and T-lineage development require the activities of E proteins. These observations have raised the question as to how E proteins influence lineage specification in both B- and T-lineage cells. We propose that in the bone marrow, E proteins by themselves in the CLP allow the initiation of a B-lineage program of gene expression, whereas in the thymus they act in concert with Notch-mediated signaling to induce a T cell fate.
Consistent with a CLP deficiency in E2A-deficient mice, we propose that E2A is transcriptionally active in the CLP cell stage (Fig. 8 A) (45). Once E2A is activated it induces the expression of a subset of genes that are essential to promote B cell development. Among the critical target genes is IL7Rα. Specifically, we show that IL-7Rα gene transcription is modulated by E47 expression. We note, however, that IL-7Rα is expressed at substantial levels in E2A-deficient HPCs, consistent with its dependence on IL-7 for cell growth (unpublished data). Thus, other factors must contribute to the regulation of IL-7Rα expression as well. One such candidate is PU.1. PU.1 has been shown to regulate IL-7Rα gene expression (46). Because PU.1 is expressed at considerable levels in E2A-deficient HPCs, it is conceivable that E proteins and PU.1 act in concert to control the expression of IL-7Rα (Fig. 8 A). IL7Rα-mediated signaling, E2A and PU.1, in turn activates EBF transcription, which in turn activates Pax5 (Fig. 8 A) (47). Once activated EBF and Pax5 then act in concert to induce a B-lineage program of gene expression (Fig. 8 A) (48). Our data also show that Bcl11a transcript levels were modestly elevated by enforced E47 expression. How Bcl11a acts in relationship to EBF and Pax5 to promote B cell development is an interesting issue that needs to be addressed.
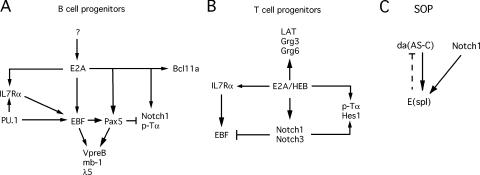
Figure 8.
Genetic regulatory networks regulating sensory organ and T and B lymphoid lineage developmental progression. Regulatory network controlling specification and commitment of B cell progenitors (A), T cell progenitors (B), and Drosophila melanogaster sensory organ progenitor (SOP) (C) are shown.
E47 also induces the expression of genes involved in the commitment of alternative cell fates, including pTα and Notch1. This brings about the question as to how the expression of these genes is antagonized in B-lineage cells. Pax5 has been shown to suppress the transcription of pTα and Notch1 (2, 49). Thus, Pax5 functions to both activate and maintain gene expression of a subset of B lineage–specific genes and, additionally, may function to repress transcription of non–B lineage–specific genes that we suggest are activated by at least in part by E2A. Collectively, we propose a genetic regulatory network in which E2A, Bcl11a, PU.1, EBF, and Pax5 act in linear and parallel pathways to initiate, repress, and maintain B lineage–specific gene transcription, promoting commitment of the CLP in the bone marrow environment to the B cell fate (Fig. 8 A).
The data demonstrate that E47 and the Notch signaling module interact at two distinct levels in fetal hematopoietic progenitors: (a) E47 acts to induce the expression of genes involved in Notch signaling, including Hes1, Hes5, Grg6, Notch3 and Notch1; (b) E47 acts in concert with Notch signaling to synergistically induce the expression of pTα and CD7 (Fig. 8 B). The induction of an ensemble of genes involved in Notch signaling by E47 is consistent with a block in thymocyte development before the onset of T-lineage commitment in E2A-deficient fetal thymocytes. We propose that this block is caused at least, in part, by an inability of these cells to receive Notch and/or integrate Notch signals. During later stages of fetal thymocyte development, E47 and Notch signaling act in a parallel pathway to promote T-lineage maturation. It will be important to determine whether similar mechanisms operate in adult thymocyte development.
Previous studies have suggested a direct link involving Notch and E2A. Notch signaling has been demonstrated to alter the transactivation activity of E47 (28, 34). Further studies have demonstrated that Notch-mediated signaling promotes the degradation of E47 upon MAP kinase–dependent phosphorylation (35, 36). Both studies suggest a linear pathway in which Notch-mediated signaling acts upstream of E-protein activity. However, we note that synergistic activation of pTα and CD7 expression by E47 and Notch signals is difficult to reconcile with a linear relationship. We note that binding sites for E47 and CSL have been identified in the regulatory elements of the pTα chain gene regulatory elements (50, 51). E47 has been demonstrated to bind directly to these enhancer elements using ChIP assays (19). How then can we reconcile a linear versus parallel relationship involving E47 and Notch-mediated signaling? Our data do not exclude the possibility that Notch signaling influences E2A activity in developing DN thymocytes. For example, it is conceivable that Notch signaling affects the transactivation activity of E47 while acting in parallel to modulate down-stream target gene expression. Likewise, it is plausible that Notch signaling in conjunction with MAP kinase signaling at the DN3 and DN4 cell stage acts to promote the degradation of E47, resulting in lower E47 proteins levels. We note that a recent report has documented that Notch signaling has the ability to overcome the block in β-selection in mice that carry mutations causing defects in pre-TCR signaling (52). Nevertheless, it will be important to assess the role of Notch signaling in modulating E2A-mediated transactivation and altered stability in the context of endogenous gene expression.
We show here that E2A proteins act to induce the expression of members of the Hes family, independent of Notch signaling. Several studies have provided evidence for a Notch-independent mechanism of Hes1 and Hes5 gene regulation. CSL-deficient mice express Hes1 and Hes5 levels in DN thymocytes that are similar to that of wild-type mice (53). Pax5-deficient pro–B cells also express substantial levels of Hes1 even in the absence of Notch signaling (43). Hes1 transcripts have been detected in fetal liver precursors, suggesting that Notch signaling was active prethymically (54). Because E47 is expressed at high levels in fetal liver progenitors, it is conceivable that Hes-1 expression in fetal liver progenitor cells is independent of Notch signaling. Thus, a role for Notch signaling in controlling Hes1 expression prethymically needs to be reevaluated. Similarly, Hes1 transcription in neuroepithelial cells is activated before the expression of Notch and Delta ligands, indicating a mechanism of regulation independent of Notch-mediate signaling (55). E47 is expressed in the neuroepithelium, and it is conceivable that also in the neuroepithelium Hes1 is regulated by E-protein activity (56).
The activation of Hes1 and Hes5 by the parallel activities of E47 and Notch signaling is intriguing because a similar regulatory network regulates Drosophila sensory organ development and notochord-specific gene expression in the ascidian Ciona intestinalis (38, 57). In flies, sensory organ development is mediated in part by Notch signaling and bHLH activity, which act in parallel to regulate the expression of the enhancer of split complex (Fig. 8 C) (38, 57). The bHLH activity in peripheral nervous system development is mediated by the proneural proteins, achaete and scute, that bind as heterodimers with daughterless, to sites present in an enhancer controlling the expression of the enhancer of split complex. We note that daughterless is closely related to the E2A proteins. Thus, the enhancer of split gene and its homologue in mammalian organisms, Hes1, are both regulated by the combined activities of Notch-mediated signaling and bHLH activity (57, 58). Notch signaling and HLH activity also regulate tissue-specific gene expression in the notochord-specific enhancer located in the Brachyuri gene of the ascidian Ciona intestinalis (57). It is plausible that an ancient basic mechanism underlying notochord-specific gene expression in ascidians and Drosophila sensory organ development has been conserved in mammalian organisms to promote T-lineage developmental progression.
MATERIALS AND METHODS
RNA purification, microarray analysis, and real-time RT-PCR.
E2A-deficient HPCs were isolated from E2A-deficient bone marrow and cultured on S17 stromal cells in the presence of SCF, IL7, and Flt3L (R&D Systems), as described previously (37). E2A-deficient HPCs were transduced with E47ER retroviral vectors as described (37). Purification of hCD25+ cells was performed using MACS beads (Miltenyi Biotec). Total RNA was isolated using an RNeasy kit (Qiagen). Microarray analysis was performed in our core facility (BIOGEM). The Codelink microarrays were used in this analysis (BD Biosciences). For real-time PCR, cDNA synthesis was performed using Superscript III (Invitrogen) following the manufacturer's protocol. Real-time PCR was performed using SYBR Green Master Mix (Stratagene) and analyzed by Mx4000 Instrumentation (Stratagene). The reactions were performed in triplicate at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s, 55°C for 1 min and 72°C for 30 s. The primer sequences used are: Hes1 forward, TTGGCTGAAAGTTACTGTGG and rev ACTATTCCAGGACCAAGGAG; Hes5 forward TACCTGAAACACAGCAAAGC and reverse GCTGGAGTGGTAAGCAG; Notch1 forward ACAACAACGAGTGTGAGTCC and reverse ACACGTGGCTCCTGTATATG; Notch2 forward TGACTGTTCCCTCACTATGG and reverse CACGTCTTGCTATTCCTCTG; Notch3 forward AGATCAATGAGTGTGCATCC and reverse GCAGACTCCATGACTACAGG; NumbL forward TTCGAGATTGAACTGTAGCC and reverse AGTGAAATGGTTCCCTTAGC; Grg6 forward GATGACAACATTTGGACAGG and reverse TAAGATGGTGTTGTCCTTGG; Grg3 forward TGAGAAGAACCACCATGAAC and reverse TGAGAAGAACCACCATGAAC; p-Tα forward CTGCAACTGGGTCATGCTTC and reverse TCAGAGGGGTGGGTAAGATC; IL-7Rα forward TTACTTCAAAGGCTTCTGGAG and reverse CTGGCTTCAACGCCTTTCACCTCA; LAT forward AGAATGTGGATGCAGATGAG and reverse CGTAATCTTCACACGACTCC; ARP forward CGACCTGGAAGTCCAACTAC and reverse ATCTGCTGCATCTGCTTG.
OP9 cocultures and flow cytometric analysis.
OP9-GFP or OP9-DL1 stromal cells were cultured in minimum essential medium alpha (MEMα; Invitrogen), supplemented with 10% fetal bovine serum (Gemini), 100 U/ml penicillin, 100 μg/ml streptomycin, and l-glutamine (Invitrogen), and plated 1 or 2 d before use in a 24-well plate, as described (20). The differentiation medium consisted of MEMα supplemented with 10% FCS (Gemini), 50 μM 2-ME, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, 5 ng/ml IL-7, and 5 ng/ml Flt3L. Lineage marker (Lin)− progenitors isolated from E2A+/+, E2A+/−, and E2A−/− fetal liver by MACS purification were seeded in 1 ml of IL-7/Flt3L medium at 105 cells/well of a 24-well plate. Various concentration of γ-secretase inhibitor IX (Calbiochem) was added to the culture, as indicated (Fig. 6). At day 7 of culture, cells were harvested and analyzed by flow cytometry. FACS analyses were performed as described (37). All antibodies were purchased from BD Biosciences or eBioscience.
Retroviral transduction and fetal thymic organ culture.
Control MigR1 and ICN retroviral vectors were provided by M.J. Bevan (University of Washington, Seattle, WA). Retroviral transduction and fetal thymic organ culture were performed as described (6, 37). Lin− progenitors were isolated from E2A+/+ and E2A−/− fetal liver at day 14 of gestation and transduced with MigR1 and ICN retroviral vectors. After 24 h of transduction, 104 cells were cultured with a deoxyguanosine-treated fetal thymic lobe for 17 d. The culture medium was replaced by half every 5 d. The cells were harvested and analyzed for the expression of GFP, CD3, CD4, CD8, and NK1.1.
Luciferase assay.
Luciferase reporter assay was performed as previously described (59). The 3′E-Hes1 region was amplified with the primers: forward CATGCCCCATTTCCAGGCAAG and reverse GAGCCAATTAAACAAAACAAA. E2A-deficient HPCs were used for the transfection with the various luciferase constructs.
Chromatin histone immunoprecipitation.
ChIP assays were performed essentially as described (59). 107-fixed cells from mice carrying the E2AGFP fusion protein as well as wild-type thymus were lysed in SDS lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS) on ice for 10 min. The lysates were sonicated and diluted with 9 parts of 50 mM Tris-HCl, pH 8.0, 167mM NaCl, 1.1% Triton X-100, and 0.11% sodium deoxycholate. 2 × 106 cells were used for each immunoprecipitation. Full-length A.V. polyclonal antibody (BD Biosciences) was used to label E2AGFP-DNA complexes. Immunocomplexes were bound to protein G-Sepharose beads and washed four times. The immunocomplexes were then eluted by incubating the beads at 65°C overnight in 10 mM Tris-HCl (pH 8.0), 5 mM EDTA, 300 mM NaCl, and 0.5% SDS. Eluted DNA was treated with RNase A and digested with proteinase K. DNA was purified using the PCR Purification kit (Qiagen). PCR was performed with the following primers: forward GGGTTGTCTCGGGTTTCAG and reverse CAGGGCTCGTGGTTTTCAT.
Acknowledgments
We thank Jim Posakony and members of the Murre laboratory for insightful discussions. We thank Juan Carlos Zuniga-Pflucker for OP9-GFP and OP9-DL1 cells and Yuan Zhuang for providing the E2AGFP knock-in mice. This work was supported by grants from the National Institutes of Health (to C. Muree).
The authors have no conflicting financial interests.
Abbreviations used: 4-OHT, 4-hydroxytamoxifen; ARP, acidic ribosomal protein; bHLH, basic HLH; ChIP, chromatin immunoprecipitation; CLP, common lymphoid progenitor cells; CHX, cycloheximide; DL1, delta-like 1; DN, double negative; DP, double positive; HLH, helix loop helix; HPC, hematopoietic progenitor cell; HSC, hematopoietic stem cell; MAP, mitogen-activated protein.
References
- 1.Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, G.F. Beilhack, J.A. Shimizu, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. [DOI] [PubMed] [Google Scholar]
- 2.Busslinger, M. 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22:55–79. [DOI] [PubMed] [Google Scholar]
- 3.Singh, H., K.L. Medina, and J.M. Pongubala. 2005. Contingent gene regulatory networks and B cell fate specification. Proc. Natl. Acad. Sci. USA. 102:4949–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelayo, R., R. Welner, S.S. Perry, J. Huang, Y. Baba, Y. Yokota, and P.W. Kincade. 2005. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr. Opin. Immunol. 17:100–107. [DOI] [PubMed] [Google Scholar]
- 5.Katsura, Y. 2002. Redefinition of lymphoid progenitors. Nat. Rev. Immunol. 2:127–132. [DOI] [PubMed] [Google Scholar]
- 6.Ikawa, T., K. Masuda, M. Lu, N. Minato, Y. Katsura, and H. Kawamoto. 2004. Identification of the earliest prethimic T-cell progenitors in murine fetal blood. Blood. 103:530–537. [DOI] [PubMed] [Google Scholar]
- 7.Masuda, K., M. Itoi, T. Amagai, N. Minato, Y. Katsura, and H. Kawamoto. 2005. Thymic anlage is colonized by progenitors restricted to T, NK and dendritic cell lineages. J. Immunol. 174:2525–2532. [DOI] [PubMed] [Google Scholar]
- 8.Allman, D., A. Sambandam, S. Kim, J.P. Miller, A. Pagan, D. Well, A. Meraz, and A. Bandoola. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174. [DOI] [PubMed] [Google Scholar]
- 9.Porritt, H.E., L.L. Rumfelt, S. Tabrizifard, T.M. Schmitt, J.C. Zuniga-Pflucker, and H.T. Petrie. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 20:735–745. [DOI] [PubMed] [Google Scholar]
- 10.Rothenberg, E.V., and T. Taghon. 2005. Molecular genetics of T cell development. Annu. Rev. Immunol. 23:601–649. [DOI] [PubMed] [Google Scholar]
- 11.Murre, C., P.S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer, daughterless, MyoD and myc proteins. Cell. 56:777–783. [DOI] [PubMed] [Google Scholar]
- 12.Murre, C. 2005. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6:1079–1086. [DOI] [PubMed] [Google Scholar]
- 13.Sawada, S., and D.R. Littman. 1993. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol. Cell. Biol. 13:5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bain, G., E. Robanus Maandag, D. Izon, D. Armsen, A. Kruisbeek, B.C. Weintraub, I. Krop, M.S. Schlissel, A. Feeney, M. van Roon, et al. 1997. E2A deficient mice show abnormalities in αβ T cell development and rapidly develop malignant T cell lymphomas. Mol. Cell. Biol. 17:4782–4791. 9234734 [Google Scholar]
- 15.Heemskerk, M.H.M., B. Blom, G. Nolan, A.P.A. Stegman, A.Q. Bakker, K. Weijer, P.C.M. Res, and H. Spits. 1997. Inhibition of T cells and promotion of natural killer cell development by the dominant negative helix loop helix factor Id 3. J. Exp. Med. 186:1597–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barndt, R.J., M. Dai, and Y. Zhuang. 2000. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell. Biol. 20:6677–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, I., C. John, G. Bain, R.R. Rivera, and C. Murre. 2001. Early thymocyte development is regulated by modification of E2A protein activity. J. Exp. Med. 194:733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlissel, M., A. Voronova, and D. Baltimore. 1991. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 5:1367–1376. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay, M., S. Herblot, E. Lecuyer, and T. Hoang. 2003. Regulation of pTα gene expression by a dosage of E2A, HEB and SCL. J. Biol. Chem. 278:12680–12687. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt, T.M., and J.C. Zuniga-Pflucker. 2002. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 17:749–756. [DOI] [PubMed] [Google Scholar]
- 21.Guidos, C.J. 2002. Notch signaling in lymphocyte development. Semin. Immunol. 14:395–404. [DOI] [PubMed] [Google Scholar]
- 22.Robey, E.A., and J.A. Bluestone. 2004. Notch signaling in lymphocyte development and function. Curr. Opin. Immunol. 16:360–366. [DOI] [PubMed] [Google Scholar]
- 23.Radtke, F., A. Wilson, S.J.C. Mancini, and H.R. Macdonald. 2004. Notch regulation of lymphocyte development and function. Nat. Immunol. 5:247–253. [DOI] [PubMed] [Google Scholar]
- 24.Maillard, I., T. Fang, and W.S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945–974. [DOI] [PubMed] [Google Scholar]
- 25.Taghon, T.N., E.S. David, J.C. Zuniga-Pflucker, and E.V. Rothenberg. 2005. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 19:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghon, T., M.A. Yui, R. Pant, R.A. Diamond, and E.V. Rothenberg. 2006. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 24:53–64. [DOI] [PubMed] [Google Scholar]
- 27.Radtke, F., A. Wilson, G. Stark, J. Bauer, V. Meerwijk, H.R. Macdonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch 1. Immunity. 10:547–588. [DOI] [PubMed] [Google Scholar]
- 28.Pui, J.C., D. Allman, L. Xu, S. DeRocco, F.G. Karnel, S. Bakkour, J.Y. Lee, T. Kadesch, R.R. Hardy, J.C. Aster, and W.S. Pear. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. [DOI] [PubMed] [Google Scholar]
- 29.Koch, U., T.A. Lcombe, D. Holland, J.L. Bowman, B.L. Cohen, S.E. Egan, and C.J. Guidos. 2001. Subversion of the T/B lineage decision in the thymus by Lunatic Fringe-mediated inhibition of Notch-1. Immunity. 15:225–236. [DOI] [PubMed] [Google Scholar]
- 30.Han, H., K. Tanigaki, N. Yamamoto, K. Kurora, M. Yoshimoto, T. Nakahata, K. Ikuta, and T. Honjo. 2002. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14:637–645. [DOI] [PubMed] [Google Scholar]
- 31.Izon, D.J., J.C. Aster, Y. He, A. Weng, F.G. Karnel, V. Patriub, L. Xu, S. Ballour, C. Rodriguez, D. Allman, and W.S. Pear. 2002. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch 1. Immunity. 16:231–243. [DOI] [PubMed] [Google Scholar]
- 32.Yun, T.J., and M.J. Bevan. 2003. Notch-regulated Ankyrin-repeat protein inhibits Notch1 signaling pathways involved in T cell development. J. Immunol. 170:5834–5841. [DOI] [PubMed] [Google Scholar]
- 33.Tomita, K., M. Hattori, E. Nakamura, E. Nakanishi, N. Minato, and R. Kageyama. 1999. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ordentlich, P., A. Lin, C.H. Shen, C. Blaumueller, K. Matsuno, S. Artavanis-Tsakonas, and T. Kadesch. 1998. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol. 18:2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nie, L., M. Xu, A. Vladimirova, and X.H. Sun. 2003. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 22:5780–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang, Z., L. Nie, M. Xu, and X.H. Sun. 2004. Notch-induced E2A degradation requires CHIP and Hsc70 as novel facilitators of ubiquitination. Mol. Cell. Biol. 24:8951–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikawa, T., H. Kawamoto, L.Y.T. Wright, and C. Murre. 2004. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 20:349–360. [DOI] [PubMed] [Google Scholar]
- 38.Bailey, A.M., and J.W. Posakony. 1995. Suppressor hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9:2609–2622. [DOI] [PubMed] [Google Scholar]
- 39.Van Doren, M., H.M. Ellis, and J.W. Posakony. 1991. The Drosophila extramacrochaete protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 113:245–255. [DOI] [PubMed] [Google Scholar]
- 40.Tabrizifard, S., A. Olaru, J. Plotkin, M. Fallahi-Sichani, F. Livak, and H.T. Petrie. 2004. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J. Immunol. 173:1094–1102. [DOI] [PubMed] [Google Scholar]
- 41.Tan, J.B., I. Visan, J.S. Yuan, and C.J. Guidos. 2005. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat. Immunol. 6:671–679. [DOI] [PubMed] [Google Scholar]
- 42.Zhuang, Y., L. Jackson, A. Pan, K. Shen, and M. Dai. 2004. Regulation of E2A gene expression in B-lymphocyte development. Mol. Immunol. 40:1165–1177. [DOI] [PubMed] [Google Scholar]
- 43.Hofflinger, S., K. Kesavan, M. Fuza, C. Hutter, B. Heavy, F. Radtke, and M. Busslinger. 2004. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 173:3935–3944. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt, T.M., M. Ciofani, H.T. Petrie, and J.C. Zuniga-Pflucker. 2004. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J. Exp. Med. 200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borghesi, L., J. Aites, S. Nelson, P. Lefterpv, P. James, and R. Gerstein. 2005. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J. Exp. Med. 202:1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeKoter, R.P., H.J. Lee, and H. Singh. 2002. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 16:297–309. [DOI] [PubMed] [Google Scholar]
- 47.Kikuchi, K., A.Y. Lai, C.L. Hsu, and M. Kondo. 2005. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 201:1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maier, H., R. Ostraat, H. Gao, S. Fields, S.A. Shinton, K.L. Medina, T. Ikawa, C. Murre, H. Singh, R.R. Hardy, and J. Hagman. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat. Immunol. 5:1069–1077. [DOI] [PubMed] [Google Scholar]
- 49.Nutt, S.L., B. Heavey, A.G. Rolink, and M. Busslinger. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax-5. Nature. 401:556–562. [DOI] [PubMed] [Google Scholar]
- 50.Reizis, B., and P. Leder. 2001. The upstream enhancer is necessary and sufficient for the expression of the pre-T cell receptor α gene is immature T lymphocyte. J. Exp. Med. 194:979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reizis, B., and P. Leder. 2002. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 16:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campese, A.F., A.I. Garbe, F. Zhang, F. Grassi, I. Screpanti, and H. von Boehmer. 2006. Notch1-dependent lymphomagenesis is assisted by but does not essentially require pre-TCR signaling. Blood. In press. [DOI] [PMC free article] [PubMed]
- 53.Tanigaki, K., M. Tsuji, N. Yamamoto, H. Han, J. Tsukada, H. Inoue, M. Kubo, and T. Honjo. 2004. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 20:611–622. [DOI] [PubMed] [Google Scholar]
- 54.Harman, B.C., W.E. Jenkinson, S.M. Parnell, S.W. Rossi, E.J. Jenkinson, and G. Anderson. 2005. T/B lineage choice occurs prior to intrathymic Notch signaling. Blood. 106:886–892. [DOI] [PubMed] [Google Scholar]
- 55.Kageyama, R., T. Ohtsuka, J. Hatakeyama, and R. Ohsawa. 2005. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 306:343–348. [DOI] [PubMed] [Google Scholar]
- 56.Roberts, V.J., R. Steenbergen, and C. Murre. 1993. Localization of E2A mRNA expression in developing and adult rat tissues. Proc. Natl. Acad. Sci. USA. 90:7583–7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corbo, J.C., S. Fujiwara, M. Levine, and A. Di Gregorio. 1998. Suppressor of hairless activates Brachyuri expression in the Ciona embryo. Dev. Biol. 203:358–368. [DOI] [PubMed] [Google Scholar]
- 58.Nellesen, D.T., E.C. Lai, and J.W. Posakony. 1999. Discrete enhancer elements mediate selective responsiveness of Enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213:33–53. [DOI] [PubMed] [Google Scholar]
- 59.Sayegh, C.E., M.W. Quong, Y. Agata, and C. Murre. 2003. E-proteins directly regulate activation induced deaminase gene expression in mature-B cells. Nat. Immunol. 4:586–594. [DOI] [PubMed] [Google Scholar]