Abstract
Celiac disease is an intestinal inflammatory disorder induced by dietary gluten in genetically susceptible individuals. The mechanisms underlying the massive expansion of interferon γ–producing intraepithelial cytotoxic T lymphocytes (CTLs) and the destruction of the epithelial cells lining the small intestine of celiac patients have remained elusive. We report massive oligoclonal expansions of intraepithelial CTLs that exhibit a profound genetic reprogramming of natural killer (NK) functions. These CTLs aberrantly expressed cytolytic NK lineage receptors, such as NKG2C, NKp44, and NKp46, which associate with adaptor molecules bearing immunoreceptor tyrosine-based activation motifs and induce ZAP-70 phosphorylation, cytokine secretion, and proliferation independently of T cell receptor signaling. This NK transformation of CTLs may underlie both the self-perpetuating, gluten-independent tissue damage and the uncontrolled CTL expansion leading to malignant lymphomas in severe forms of celiac disease. Because similar changes were detected in a subset of CTLs from cytomegalovirus-seropositive patients, we suggest that a stepwise transformation of CTLs into NK-like cells may underlie immunopathology in various chronic infectious and inflammatory diseases.
Tissue epithelial cells actively signal the presence or absence of stress through the induction of cytokines and surface ligands, which, in turn, engage coactivating or inhibitory NK receptors expressed by effector CTLs (for review see reference 1). One prominent pathway in this crosstalk between the tissue target and the effector CTLs involves NKG2D (2–6) and its multiple ligands: MICA/B (7) and ULBPs (8) in humans and Rae 1α-ɛ, H60, and MULT1 in mice (9–11). NKG2D exclusively associates in humans (4, 12–14), in contrast to mice (15, 16), with the PI3 kinase–bearing adaptor molecule DAP10 (4, 12–14), which endows NKG2D with costimulatory properties for TCR activation (2, 7).
However, we recently reported that preexposure to IL-15, a cytokine up-regulated in inflammatory and infectious conditions (17), enabled NKG2D to selectively unleash the cytolytic properties of effector CTLs, independently of TCR specificity (2, 4). The arming of NKG2D by IL-15 to induce cytolysis in CTLs may be particularly relevant in the pathogenesis of T cell–mediated diseases, where IL-15 is up-regulated by tissue target cells (for review see reference 1). A good example is celiac disease, where DQ2- and DQ8-restricted CD4+ T cell responses in the lamina propria to peptides derived from dietary gluten are necessary (for review see references 18 and 19) but not sufficient (20) to induce villous atrophy and malabsorption, the hallmarks of celiac disease (for review see references 21–23). Despite the dramatic expansion of intraepithelial CTLs (IE-CTLs) in active celiac (AC) disease, their role was for a long time disregarded because gluten-specific IE-CTLs could not be identified (for review see references 21 and 22). A potential molecular basis for epithelial cell destruction by IE-CTLs and the slow and partial recovery of a normal mucosa after gluten exclusion (24, 25) was provided by the finding that NKG2D expressed on IE-CTLs mediated cytolysis of stressed MICA/B-expressing enterocytes (4, 5).
However, because NKG2D could not induce other effector functions, such as cytokine secretion and proliferation in IE-CTLs (2, 4), several findings remained obscure. First, it was shown in AC disease that IE-CTLs secreted high levels of IFN-γ (26, 27). Second, a long-term complication of celiac disease is “refractory sprue,” a severe diet-refractory condition associated with a massive infiltration of the diseased epithelium by CTLs, which ultimately undergo malignant transformation into lymphomas (for review see references 21, 28, and 29). Collectively, these observations led us to hypothesize that IE-CTLs, in an intestinal environment favoring their chronic activation, may have undergone a dysregulation of their genetic program, resulting in the aberrant expression of NK receptors associated with immunoreceptor tyrosine-based activation motif (ITAM)-bearing adaptor molecules capable of mediating proliferation and cytokine secretion.
Here, we identify in celiac patients a massive expansion of a few IE-CTL clones that have undergone a genetic reprogramming of their signaling properties, which has essentially converted them into functional NK cells. The transcriptional signature of this reprogramming is the induction of a panoply of receptors and adapters normally restricted to the NK lineage. One conspicuous example is CD94/NKG2C, which alone signals cell proliferation, cytokine secretion, and target killing through the ITAM-bearing adaptor molecule DAP12 (30), also named KARAP (31), without a requirement for TCR engagement. In addition, its ligand HLA-E (32, 33) is strongly induced on celiac enterocytes, hence enabling full activation of intraepithelial NKG2C+ CTLs.
This broad acquisition of the NK signaling program in CTLs may significantly contribute to celiac intestinal immunopathology, as it violates the well-established rule of cell-mediated immunity indicating that CTLs may not be able to proliferate, produce inflammatory cytokines, or kill unless their TCR specifically recognizes antigen presented by MHC class I molecules on the target cell membrane.
RESULTS
Selective expansion of CTLs expressing activating CD94/NKG2C receptors in the intestinal epithelium of celiac patients
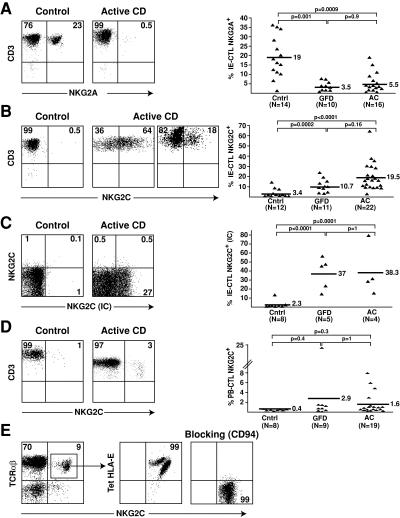
CD94/NKG2C receptors, which are classically absent in CTLs, were reported to be expressed in CMV-seropositive patients on a small subset of human CTLs of unknown specificity (34, 35). In the normal intestinal epithelium, CD94 is expressed by a subset of IE-CTLs and associates predominantly with the inhibitory NKG2A subunit (36). Previous studies have reported a marked expansion of CD94+ IE-CTLs in celiac disease (37). In this study, we sought to determine the nature, inhibitory or activating, of the NKG2 subunit associated with CD94. Fig. 1 A shows that the inhibitory CD94/NKG2A receptor was markedly underrepresented. Thus, whereas ∼20% (n = 14; mean: 19 ± 11%) of healthy IE-CTLs expressed NKG2A, less than 6% of their celiac counterparts did, whether active (n = 16; mean: 5.5 ± 5.5%) or on a gluten-free diet (GFD; n = 10; mean: 3.5 ± 2.5%). In contrast, surface NKG2C, which was normally expressed at a very low frequency on healthy IE-CTLs (n = 12; mean: 3.4 ± 4.4%), was found on the cell surface of a substantial proportion of IE-CTLs from AC patients (n = 22; mean: 19.5 ± 13.8%; P < 0.0001; Fig. 1 B). In 7 out of 22 patients, the frequency of NKG2C+ IE-CTLs was >20% and was 64% in one patient (Fig. 1 B). Notably, patients on GFD exhibited a decrease, albeit not significant, in the frequency of NKG2C+ IE-CTLs (n = 11; mean: 10.7 ± 6.5%; P = 0.16), suggesting that NKG2C expression remained increased after exclusion of gluten (Fig. 1 B). Importantly, NKG2A, when present, was never coexpressed with NKG2C, as reported previously (36, 38).
Figure 1.
Selective expansion of IE-CTLs expressing activating CD94/NKG2C receptors in celiac disease. Representative flow cytometric data on freshly isolated TCR-αβ+CD8+ IE-CTLs or PB-CTLs from normal controls (Cntrl), AC patients, and celiac patients on a GFD are shown in the right panels. A scatter plot graph summarizing all samples investigated is shown on the right in A–D. (A) Decreased frequency of IE-CTLs expressing inhibitory CD94/NKG2A receptors in AC patients and GFD patients. (B) Increased frequency of IE-CTLs expressing surface CD94/NKG2C receptors in AC patients and GFD compared with controls. (C) Increased frequency of IE-CTLs expressing intracellular NKG2C protein (NKG2C IC) in AC patients and GFD compared with controls. (D) Propensity toward an increased frequency of PB-CTLs expressing surface CD94/NKG2C receptors in AC patients and GFD compared with controls. (E) CD94-dependent HLA-E recognition by CD94/NKG2C+ CTLs. A representative example of HLA-E staining on gated TCR-αβ+CD8+ NKG2C+ CTLs is shown on freshly isolated PBLs. Left panel shows that incubation with anti-CD94 antibody before incubation with a HLA-E tetramer prevents staining with the HLA-E tetramer.
CD94/NKG2C is a heterodimeric receptor (for review see reference 39) whose surface expression depends on the expression of the signaling adaptor DAP12 (30). Because DAP12 expression might be regulated independently of NKG2C, we also stained NKG2C after membrane permeabilization to detect intracellular NKG2C (Fig. 1 C). On average, nearly 40% of the permeabilized IE-CTLs from either patients with AC disease (n = 4; mean: 38.3 ± 28%; P = 0.0001) or on a GFD (n = 5; mean: 37 ± 17.7%; P < 0.0001) expressed NKG2C, whereas IE-CTLs from healthy controls did not (n = 8; mean: 2.3 ± 4.4%). This surprising finding revealed a more profound dysregulation of NKG2C than anticipated by analysis of NKG2C on the cell surface of IE-CTLs, even in apparently symptom-free GFD patients.
Among the peripheral blood CD8+TCR-αβ+ of healthy individuals (peripheral blood CTLs [PB-CTLs]), CD94/NKG2C+ CTLs were rarely detected (n = 8; mean: 0.4 ± 0.1%), as reported previously (35). On average, the frequency in celiac samples (n = 19; mean: 1.6 ± 2.2%; P = 0.3) was not statistically different from healthy samples, although a few patients did exhibit unusual frequencies as high as 5–10% (Fig. 1 D).
To further characterize CD94/NKG2C expression on celiac CTLs, we used tetramers of its ligand, HLA-E, loaded with an HLA-G leader peptide and demonstrated staining of 99% of the NKG2C+ CTLs, as expected (Fig. 1 E). Interestingly, however, the example shown in Fig. 1 E indicated that two subsets of NKG2C+ HLA-E tetramer+ CTLs were resolved based on the staining intensity with anti-NKG2C antibody and HLA-E tetramers, respectively. The nature of these subsets is unclear, but the data might indicate the presence of another HLA-E binding CD94/NKG2 isoform, e.g., NKG2H or NKG2E, coexpressed with NKG2C (40). Tetramer staining was blocked by an anti-CD94 antibody, indicating that HLA-E recognition was exclusively mediated by CD94-bearing receptors.
Collectively, these results indicate that there is a selective up-regulation of activating CD94/NKG2C receptors on celiac IE-CTLs, potentially tipping the balance toward activation of CTL effector function. Furthermore, our results suggest that expression of NKG2C persists partially aftergluten exclusion.
CTL stimulation by NKG2C/DAP12 and costimulation by NKG2D
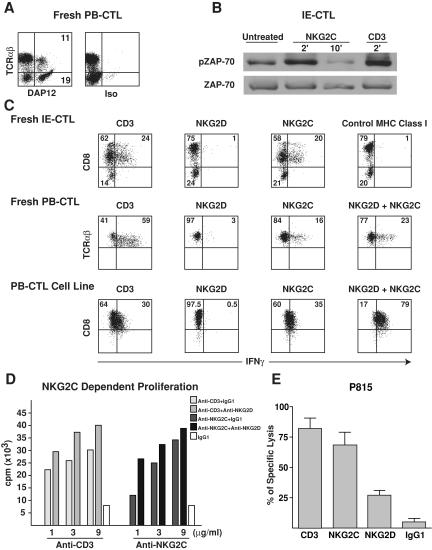
Because DAP12 is essential for CD94/NKG2C expression at the cell surface (30), we investigated whether DAP12 was present and formed a functional complex with CD94/NKG2C in NKG2C+ IE-CTLs and PB-CTLs. As anticipated, DAP12 transcripts and protein were absent in normal IE-CTLs (not depicted). In contrast, DAP12 was expressed in five out of five NKG2C+ CTLs examined. A representative intracellular DAP12 staining is shown in the PB-CTLs of a patient with AC disease (Fig. 2 A). DAP12 was expressed at comparable levels in TCR-αβ+ T cells and in NK cells (TCR-αβ−). Furthermore, surface coimmunoprecipitation experiments performed on celiac IE-CTL clones that expressed both CD94/NKG2C and NKG2D showed that NKG2C was associated with DAP12, whereas NKG2D was associated with DAP10 (reference 4 and not depicted). Finally, stimulation of the IE-CTL clone by anti-NKG2C induced ZAP-70 phosphorylation (Fig. 2 B), as established previously with NK cells (41).
Figure 2.
Expression of functional CD94/NKG2C/DAP12 immunoreceptor complexes by celiac CTLs. (A) A representative intracellular DAP12 staining in TCR-αβ T cells is shown on celiac PB-CTLs. Similar results were obtained with freshly purified celiac NKG2C+ IE-CTLs and IE-CTL clones as well as NKG2C+ PB-CTLs from a CMV-seropositive individual. (B) Ligation of NKG2C, similarly to CD3, induced ZAP-70 phosphorylation in NKG2C+ IE-CTLs. Staining with total anti–ZAP-70 mAb demonstrates equal loading. (C and D) NKG2D costimulates NKG2C-mediated cytokine secretion and proliferation. Fresh NKG2C+ CTLs and NKG2C+ CTL lines and clones were incubated with plate-bound mAb, and IFN-γ secretion (C) and proliferation (D) were determined by flow cytometry or [3H]thymidine uptake, respectively. Experiments were performed in triplicate, and error bars were <10%. Representative examples of three independent experiments are shown. Of interest, CD8 was down-regulated upon TCR and NKG2C, but not NKG2D, stimulation in freshly isolated IE-CTLs. (E) Anti-NKG2C, similarly to anti-CD3 cross-linking, induced redirected lysis of P815 target cells by NKG2C+ IE-CTL and PB-CTL clones. In all the clones tested, NKG2C was always more efficient in inducing lysis of P815 targets than NKG2D. In contrast, cross-linking with isotype-matched control mAb was ineffective. A representative experiment using four NKG2C+ IE-CTL clones from the same patient is shown.
In a plate-bound antibody stimulation assay, NKG2C engagement induced IFN-γ secretion (Fig. 2 C) as well as proliferation (Fig. 2 D) of celiac NKG2C+ CTLs. Notably, although NKG2D did not induce cytokine secretion and proliferation by itself, it did enhance NKG2C-mediated stimulation (Fig. 2, C, right panels, and D), thus acting as a costimulatory receptor for NKG2C. As reported previously, NKG2D induced cytolysis of the Fcγ+ P815 mastocytoma cell line in a mAb-redirected cytotoxic assay (4), albeit consistently to a lower level than NKG2C (Fig. 2 E).
These findings demonstrate that NKG2C can substitute for the TCR to activate celiac CTLs, and that NKG2D costimulates NKG2C-mediated cytokine secretion and proliferation.
HLA-E up-regulation by celiac enterocytes
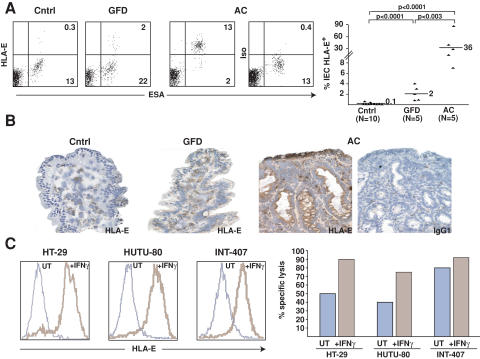
Interestingly, HLA-E, similarly to MICA/B (4, 5), is not expressed on the surface of enterocytes in the small bowel of normal individuals (n = 10; mean: 0.1 ± 0.2%; P < 0.0001; Fig. 3 A). In contrast, flow cytometric analysis revealed that HLA-E was massively induced on freshly isolated enterocytes of AC patients (n = 5; mean: 36 ± 32.2%), reaching up to 90% in one patient. Importantly, HLA-E was down-regulated in patients after initiation of a GFD (n = 5; mean: 2.1 ± 1.4%; P < 0.003), similarly to the previously reported down-regulation of MICA/B (4, 5). By immunohistochemistry, HLA-E was found to be most highly expressed in the enterocytes lining the atrophied villi of celiac patients (Fig. 3 B). One major inducer of HLA-E is IFN-γ (42), which is highly expressed in the intestinal mucosa of AC patients. Indeed, IFN-γ induced HLA-E surface expression in three out of three enterocyte lines tested (Fig. 3 C). Consistent with these observations, preexposure of enterocyte cell lines to IFN-γ sensitized them for killing by celiac NKG2C+ CTL lines. One apparent exception was INT-407 (Fig. 3 C), which was shown to be efficiently killed by NKG2D+ (NKG2C−) IE-CTLs (4).
Figure 3.
HLA-E up-regulation in celiac enterocytes and killing of HLA-E+ enterocytes by celiac CTLs. (A) Flow cytometric analysis of freshly isolated enterocytes (ESA+) from AC patients shows surface HLA-E expression. The scatter plot summarizes the percentage of HLA-E+ enterocytes in control (Cntrl), GFD, and AC patients. (B) Immunohistochemical analysis of HLA-E expression in enterocytes of AC, GFD and control patients. A representative example out of five is shown. (C) HLA-E expression by enterocytes cultured overnight in medium alone (UT) or with IFN-γ was determined by flow cytometry using anti–HLA-E mAb (brown line) or isotype control (blue line). Note that HLA-E was expressed significantly only after stimulation with IFN-γ in the three enterocyte lines tested. Interestingly, specific killing of 51Cr-labeled enterocyte lines by IE-CTL lines was increased when enterocytes were preincubated with IFN-γ.
Collectively, these data indicate that the expression of functional CD94/NKG2C receptors by celiac IE-CTLs is met by a corresponding induction of their HLA-E ligand on enterocytes, likely contributed in part by increased IFN-γ levels in the gut mucosa.
Global NK reprogramming of NKG2C+ celiac CTLs
CD94/NKG2C/DAP12 immunoreceptor complexes are normally not expressed in CTLs. Therefore, we hypothesized that their expression might be the reflection of a more general and profound dysregulation of genes related to the NK lineage. To test this hypothesis, we analyzed the transcriptional phenotype of NKG2C+ CTLs using a microarray approach in combination with flow cytometric and functional analysis.
We first performed a microarray analysis for gene expression on NKG2C+ and NKG2C− CTLs. IE-CTL (n = 8) and PB-CTL (n = 4) clones were derived from four celiac patients. For each of the two IE-CTL samples, two NKG2C+ and two NKG2C− clones were analyzed, and the mean gene expression level was taken into account for the analysis. In addition, one pair of NKG2C+ and NKG2C− CTL clones derived from a CMV-seropositive patient (patient 2) was examined, as the presence of functional CD94/NKG2C/DAP12 receptors was recently reported on the CTLs of these individuals (34, 35). The clones were derived from sorted single NKG2C+ and NKG2C− cells after only one round of expansion (corresponding to 15–18 d of cell culture) to minimize changes associated with long-term cultures. To exclude genetic heterogeneity between individuals, comparisons were done in a pairwise manner between NKG2C+ and NKG2C− clones originating from the same individuals. Two types of analysis were completed. A global analysis (Fig. 4 A) comparing all NKG2C+ and NKG2C− clones was performed to identify genes that were consistently differentially expressed by NKG2C+ and NKG2C− CTLs, regardless of their origin. In addition, a second analysis was performed on celiac IE-CTLs and PB-CTLs separately (the CMV-seropositive sample was excluded) to allow a more specific analysis of individual cell types and reveal additional differences masked by the global analysis (Table S1, available at http://www.jem.org/cgi/content/full/jem.20060028/DC1).
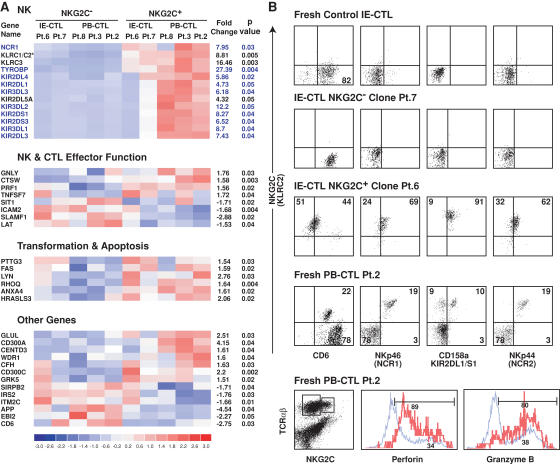
Figure 4.
NK reprogramming in NKG2C+ CTLs. (A) The 40 significant genes obtained by global pairwise microarray analysis of NKG2C+ and NKG2C− IE-CTL and PB-CTL clones generated from four celiac patients and one CMV-seropositive patient are illustrated. 27 of the 40 genes could be clustered into three distinct groups based on their associated function. The remaining 13 genes are listed under “Other Genes.” On the left hand side of the cluster analysis is the gene name as designated by HGNC. An asterisk (*) indicates the inability of the GeneChip to distinguish between the two genes (KLRC1 and KLRC2). However, based on data presented in Fig. 1 and analysis of NKG2A and NKG2C transcripts by RT-PCR using specific primers, this gene was identified as KLRC2 (NKG2C). On the right hand side of the cluster analysis, the fold change between the NKG2C+ and NKG2C− populations is shown along with the p-value for a paired t test. Of interest, 10 out of 13 genes in the NK cluster (highlighted in blue) are all located within the NK receptor cluster on chromosome 19q13. (B) This panel highlights the phenotypic correlation of the microarray data by multicolor flow cytometry. Representative dot plots of NKp46 (NCR1), CD158a (KIR2DL1/KIR2DS1), and CD6 expression gated on TCR-αβ+ NKG2C+ populations from fresh normal IE-CTLs, NKG2C+ and NKG2C− IE-CTL clones, and fresh PB-CTLs from a CMV-seropositive patient are shown. Because CD6 expression in the NKG2C− CTL subset of patient 2 (Pt.2) was high, the compensation criteria were distinct from those used with the NK receptors NKp46, CD158a, and NKp44. This resulted in a lower level of NKG2C staining when anti-NKG2C antibody was associated with anti-CD6 antibody. Interestingly, NKG2C+ but not NKG2C− CTLs also express NKp44 (NCR2), a result that was not anticipated by microarray analysis. In addition, NKG2C+ CTLs (in red) show increased expression of both perforin and granzyme B compared with NKG2C− CTLs (in blue), as suggested by the microarray analysis.
Strikingly, the global analysis identified only 40 genes that were consistently differentially expressed by NKG2C+ and NKG2C− CTLs. Among these 40 genes, 27 could be included into three functionally relevant groups: “NK,” “NK and CTL effector function,” and “transformation and apoptosis.” The remaining 13 genes were classified as “other genes.” Of interest, among the 40 significant genes identified in this study, as many as 19 were related to NK cell phenotype and function, a very significant finding (P < 0.0001), highlighting the existence of a specific NK transcriptional program associated with NKG2C. As expected, expression levels of KLRC2 (NKG2C) and TYROBP (DAP12) genes were significantly increased (8.81-fold change, P = 0.005 and 27.39-fold change, P = 0.004, respectively) in NKG2C+ CTLs. In addition, we found that a much broader set of NK receptor genes was increased in NKG2C+ CTLs. These included the C-type lectin receptor NKG2E (KLRC3) and the NCR1 receptor NKp46. A very large set of NK receptors of the KIR family was also up-regulated, usually at higher levels in PB-CTLs than in IE-CTLs (see Fig. 4 A and Table S1). dChip analysis indicated that genes differentially expressed between the NKG2C+ and NKG2C− subsets were preferentially localized to chromosome 19q13, which comprises the NK cluster encoding immunoglobulin-like NK receptors and the NCR NKp46 receptor (P < 0.001; illustrated in Fig. 4 A in blue). Finally, NKG2C+ CTLs also had increased expression of genes related to cytolytic effector functions, including granulysin, cathepsin W, and perforin 1.
The individual analysis (Table S1) confirmed the global microarray analysis and revealed the up-regulation of additional NK-related genes. In addition, it suggested a significant decrease in the transcript levels of both the TCR-α (1.56-fold) and TCR-β (3.28-fold) chains (Table S1). This latter observation was confirmed in eight out of eight clones by quantitative RT-PCR, which showed a 4.8-fold decrease in the TCR-β transcripts of NKG2C+ (mean: 0.32 ± 0.44; range: 0.14–0.55) compared with control NKG2C− IE-CTLs (mean: 1.53 ± 0.45; range: 0.97–1.93). This intriguing finding may relate to the NK reprogramming and is consistent with the down-regulation of TCR by transformed NK-like CTLs found in refractory celiac sprue (43).
Flow cytometric studies performed on IE-CTLs and PB-CTLs analyzed by microarray confirmed, at the protein level, the expression of additional NK receptors and the up-regulation of granzyme and perforin in NKG2C+ CTLs (Fig. 4 B). Flow cytometric dot plots of representative molecules that were reported to be significantly down-regulated (CD6) or up-regulated (NKp46 [NCR1] and CD158a [KIR2DL1/S1]) in the global microarray analysis are shown (Fig. 4 B). Flow cytometric data on freshly isolated PB-CTLs from the CMV-seropositive patient (patient 2) are shown to further confirm that the NK reprogramming is not an artifact associated with cell culture. Interestingly, NKp44 expression (another NK receptor associated with DAP12; reference 44) was also induced on NKG2C+ cells (Fig. 4 B, right panels), despite RNA expression levels that were apparently similar to their NKG2C− counterpart. This observation suggests that additional posttranscriptional mechanisms are coordinated with transcriptional changes to produce the NK phenotype of NKG2C+ CTLs. This is consistent with the finding that NKG2C protein is only found in celiac IE-CTLs (Fig. 1 C), even though normal IE-CTLs and PB-CTLs express NKG2C transcripts (36). Finally, the induction of KIR2DL1/S1 (CD158a) protein on NKG2C+ CTLs in patient 6, which was not anticipated by the global microarray analysis (Fig. 4 A), indicates that the extent of induction of NK-associated genes in the NKG2C+ CTLs is probably underestimated by this global analysis, which is more specific but less sensitive. This is supported by the observation that the individual microarray analysis revealed the up-regulation of many additional NK receptors in IE-CTLs and PB-CTLs. For example, it showed an increase in additional KIRs (e.g., KIR2DL2 and S2) and CD56 in IE-CTLs, and CD16 and KLRG1 in PB-CTLs (Table S1).
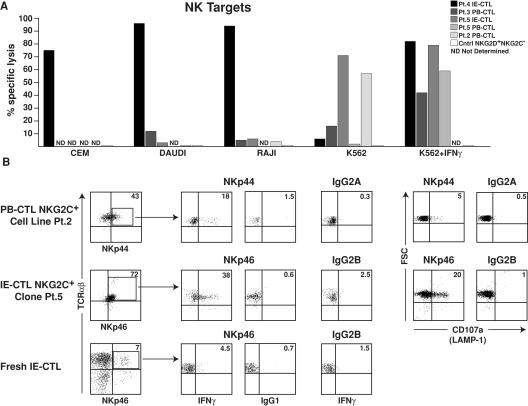
Finally, the acquisition of a NK-like phenotype by NKG2C+ CTLs was confirmed on one hand by analyzing their capacity to kill NK targets (Fig. 5) and on the other hand by demonstrating that the NK receptors of the NCR family NKp44 and NKp46 were functional. As anticipated, NKG2C+NKG2D+, but not control NKG2C− NKG2D+, celiac CTLs could efficiently kill CEM, DAUDI, RAJI, or K562 cell lines. Interestingly, IFN-γ treatment of K562 sensitized their killing by NKG2C+. The killing of IFN- γ–treated K562 and HT-29 cells could only be marginally blocked (up to 30%) by anti–HLA-E antibody, HLA-E tetramers, and anti-CD94 and NKG2D antibodies, alone or in combination (not depicted), suggesting that multiple activating NK receptors are involved in the killing of these targets. Of interest, individual NKG2C+ CTL clones exhibited vastly different cytolytic profiles against the different NK targets. These findings, similar to previous reports on NK cell clones, suggested different assortments of activating and inhibitory NK receptors on individual NKG2C+ celiac CTLs and their corresponding ligands on different targets (for review see references 45–48). Furthermore, analysis of NCR-mediated cytokine secretion and degranulation of cytolytic granules using plate-bound antibody assays revealed that NKp44 and NKp46 were not only expressed, but also functional in NKG2C+ CTLs (Fig. 5 B).
Figure 5.
NKG2C+ CTLs kill NK targets and express functional Nkp44 and NKp46 receptors. (A) Cytolytic activity of five NKG2C+ CTL clones and one NKG2C−NKG2D+ CTL clone used as control against 51Cr-labeled NK targets, CEM, DAUDI, RAJI, and K562 is shown at an E/T ratio of 50:1. Note that different NKG2C+ CTL clones, similarly to what is observed with NK cells, display distinct pattern of target cell lysis. Interestingly, IFN-γ treatment of K562 augmented their killing by NKG2C+ CTL clones. ND, not determined. Experiments were performed in triplicate, and error bars were <10%. (B) NKG2C+ PB-CTLs and IE-CTLs were incubated with 5 μg/ml plate-bound antibody or isotype control as indicated. IFN-γ production was assessed after overnight stimulation by intracellular flow cytometric analysis, and degranulation of cytolytic granules was determined by analysis of surface CD107a expression upon 4 h of stimulation.
Collectively, the phenotypic, functional, and genetic analyses of NKG2C+ CTLs indicate that they have acquired broad properties of NK cells at the genetic, phenotypic, and functional levels.
TCR repertoire analysis of NKG2C+ CTLs reveals a highly restricted repertoire
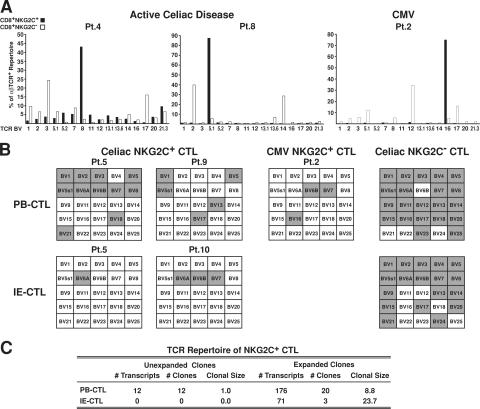
Because NKG2C has the ability to signal proliferation by itself, independently of TCR engagement, we evaluated the level of clonal expansion by examining the TCR repertoire of NKG2C+ CTLs. A combination of TCR Vβ staining, RT-PCR, and sequencing, as described previously (36), was applied to freshly isolated or short-term cultures of NKG2C+ and NKG2− CTLs from five patients with AC disease and from one CMV-seropositive individual. Fig. 6 A shows that in sharp contrast with the broad repertoire of the NKG2C− samples, a single BV family dominated the TCR repertoire in all three NKG2C+ samples. In accordance with previous studies, the TCR repertoire of NKG2C+ and NKG2C− CTLs was distinct (36, 38). The dominant BV was different in each individual: BV 8 (42%) in celiac patient 4, BV 5.1 (87%) in celiac patient 8, and BV 16 (76%) in the CMV-seropositive patient 2. TCR-β chain–specific RT-PCR confirmed and extended these results (Fig. 6 B), showing only six BV families (range: 3–11) expressed in NKG2C+ PB-CTLs compared with 21 expressed in the CD94+ NKG2C− control PB-CTL population. In addition, a very restricted number of BV families (range: 1–3) was expressed in NKG2C+ IE-CTLs compared with CD94+ NKG2C− control IE-CTLs (range: 11–18). Interestingly, comparison of the BV families between PB-CTLs and IE-CTLs suggested that no BV families were shared between the two subsets. In particular, sequence nucleotide analysis of the CDR3 region of BV 6A, the only BV family found to be expressed in both PB-CTL and IE-CTL subsets in patient 5, revealed that the BV6A T cell clones were different in the blood and the intestine. Furthermore, higher resolution analysis of the TCR repertoire of PB-CTLs and IE-CTLs expressing NKG2C using direct nucleotide sequencing of short-term cell cultures (36, 49) demonstrated that nearly all the sequences belonged to a few, yet sizeable, clonal expansions. Fig. 6 C shows that IE-CTLs and PB-CTLs had an average of 23.7 and 8.9 transcripts per clone, respectively, indicating that NKG2C+ CTLs had undergone massive oligoclonal expansions in vivo. No evidence of antigen drive was seen despite a large sequencing effort (295 TCR-β chain clonotypes generated).
Figure 6.
NKG2C+ CTLs display a highly restricted TCR repertoire. (A) TCR-β chain repertoire analysis of CD8+ NKG2C+ and NKG2C− populations from two AC disease samples and one PB-CTL sample from a CMV-seropositive patient by flow cytometry is shown. Results are expressed as percentages of TCR-β chain+ cells among CD8+TCR-αβ+ T cells. (B) TCR-β chain repertoire analysis by RT-PCR analysis of representative PB-CTL and IE-CTL samples from AC patients, PB-CTLs from a CMV-seropositive patient, and representative CD94+ NKG2C− control CTL samples matched for the effector/memory status (expression of CD45 RO and RA, CCR7, CD62L, and capacity to mediate TCR cytolysis). The TCR repertoire is represented by a grid that contains 25 separate boxes, each signifying a TCR-β chain variable region (BV) gene family (BV1-BV25). Gray shaded boxes indicate that the corresponding TCR BV gene is expressed. Patient 5 (Pt.5) has paired PB-CTL and IE-CTL samples obtained at the same time. Notably, patient 2 (Pt.2) illustrates that a similar result was obtained by both flow cytometry and RT-PCR. TCR nomenclature is designated by Arden et al. (reference 66). (C) This panel summarizes the 295 β chain nucleotide sequences obtained by sequencing 10 TCR BV families from three PB-CTL and two IE-CTL celiac samples. The CTL populations are represented by both the number of transcripts (TCR-β chain sequences) and the number of clones. Expanded clones contain two or more identical β chain sequences, whereas clonal size represents the average number of sequences per clone.
DISCUSSION
Major expansion of highly activated IFN-γ+ IE-CTLs within the atrophic intestinal epithelium is a characteristic component of celiac disease (26, 27). Although multiple studies have failed to detect gluten-specific responses among these IE-CTLs, converging reports have recently identified a dysregulation in the function of NKG2D allowing CTLs to kill stressed MICA/B+ enterocytes (4, 5). We now identify in celiac patients a population of IE-CTLs with a general and lasting alteration of NK receptor expression and function. This dysregulation involved expression of activating NK receptors, such as CD94/NKG2C, NKp44, and NKp46, considered exclusive NK lineage receptors that can directly signal through ITAM-bearing DAP12 and CD3ζ (for review see references 48, 50, and 51). The expression of these prototypic activating NK receptors endows CTLs to not only kill, but also secrete cytokines in an antigen-nonspecific manner. Furthermore, we demonstrate that NKG2C can mediate proliferation in CTLs, suggesting that proliferation is no longer under the control of the TCR.
CD94/NKG2C expression by celiac IE-CTLs appeared to be a marker for a general NK reprogramming, as attested by the significant and overall up-regulation of genes belonging to the NK cluster on chromosome 19q13 (P < 0.0001), which encodes KIRs, NKp46, and DAP12, and the observation that out of the 40 genes significantly increased in NKG2C+ CTLs, 19 were related to NK-associated molecules (P < 0.0001). These genetic alterations translated into the acquisition of a functional NK phenotype by CTLs, whereby a panoply of NK receptors could, alone or in concert, inflict damage to the host tissue. For example, NKG2D/DAP10 costimulated CD94/NKG2C/DAP12-mediated effector functions and proliferation, a finding that is particularly relevant in the context of celiac disease, where both MICA/B and HLA-E are induced on celiac enterocytes. Little is known at present about the molecular nature of the “switch” turning on this NK program in celiac effector CTLs. However, extending a recent report that PB-CTLs from CMV-seropositive individuals expressed functional CD94/NKG2C receptors (35), our detailed genetic and functional studies of one CMV-seropositive individual would indicate that several inflammatory conditions might lead to a similar NK reprogramming of CTLs. Collectively, our past (4) and present observations give a molecular basis to older studies reporting that CTL clones can be induced under particular culture conditions to exert “promiscuous,” antigen-nonspecific killing (52–54).
There are striking parallels between the existence of reprogrammed CTLs and the characteristic complications of celiac disease, refractory sprue, and enteropathy-associated T cell lymphoma (EATL). In accordance with the expression of CD94 (55) and the NK-like properties (56) reported for refractory IE-CTLs, we have found expression of functional CD94/NKG2C receptors in two out of two refractory sprue patients examined (not depicted). The TCR studies of the CD94/NKG2C+ IE-CTL subset in “uncomplicated” celiac patients, as reported here, clearly established that these reprogrammed CTLs had undergone significant clonal expansions, suggesting that by enabling CTL proliferation through TCR-independent NK receptor–mediated mechanisms, reprogramming may be the direct cause of these chronic uncontrolled expansions. Together with the dysregulation of genes associated with cell transformation (e.g., LYN and HRASLS3), these findings may suggest that the NKG2C+ IE-CTLs can ultimately undergo malignant transformation. Likewise, the reduction of TCR-α and TCR-β transcripts may represent a mechanism explaining the low or absent surface TCR expression reported in a subset of refractory sprue and EATL lymphomas (43). Future prospective studies correlating the presence of NK reprogramming with the severity of disease, the resistance to GFD, and the incidence of refractory sprue or EATL will test the predictive value of these changes. Detailed analysis of the massive clonal expansions of CD94/NKG2C+ IE-CTLs in patients developing diet-refractory disease may also lead to further insights into the transformation process.
In addition to novel insights into the pathology of celiac disease, our findings have implications on the differentiation and selection of the T cell and NK cell lineage. Indeed, although CTLs and NK cells are the two prominent lymphoid subsets in charge of cytolytic function within the vertebrate immune system, they follow fundamentally different rules for cellular activation and tolerance. Whereas CTLs use diversely rearranged TCRs to focus on rapidly evolving microbial peptides, NK cells use a limited set of germline-encoded receptors to recognize conserved stress-induced self-ligands as stimuli to rid infectious agents. CTL tolerance applies to cells bearing self-specific TCRs, whereas NK cell tolerance applies to cells lacking appropriate self-specific inhibitory receptors (57). In this context, the existence of CTL/NK hybrid cell types raises several important functional issues. For example, activating NK receptors, such as CD94/NKG2C, NKp44, and NKp46, which signal directly through ITAM-bearing DAP12, CD3ζ, or FcɛRIγ, could potentially interfere with normal T cell tolerance mechanisms by inducing the expansion and unleashing the effector functions of CTLs. Thus, several recent studies have investigated the extent of NK lineage receptor expression by subsets of effector/memory CTLs, the mechanisms of signal integration between TCR and NK receptors, and the potential consequences in health and disease. These studies have uniformly emphasized that NK lineage receptors expressed in CTLs were restricted to those serving a costimulatory and inhibitory function for TCR signaling in effector/memory CTLs, similar to the role of CD28 and CTLA-4 in naive T cells (for review see references 1 and 58). These NK receptors enhanced or prevented TCR-mediated cytolysis based on stress- or inflammation-induced expression of NK receptor ligands by the target cell. This new layer of regulation of adaptive immunity through innate signals emitted by the target itself serves a dual role of enhancing cytolysis of cells that are infected while preserving those healthy cells that might express cross-reactive self-antigens.
We speculate that the graded induction of NK lineage features and ultimately the functional transformation of CTLs into NK-like cells may represent a dysregulation of the adaptive immune response induced under conditions of chronic activation (59) and resulting in the reversal to innate modes of immune defense. Although it is advantageous to enhance the ability of CTLs to eradicate cells undergoing stress even in the presence of low or undetectable antigen, this rather indiscriminate mode of defense may ultimately lead to chronic forms of inflammation and disease. Understanding the mechanisms leading to this stepwise reprogramming of CTLs may shed new light on these chronic infectious and inflammatory diseases, as well as suggest therapies better suited to treat them.
MATERIALS AND METHODS
Patients and controls.
Diagnosis of celiac disease was based on detection of anti-transglutaminase antibodies, the presence of HLA DQ2 or DQ8, villous atrophy, and clinical and histological response to GFD. 27 individuals had AC disease with partial villous atrophy, whereas 16 were on a strict GFD for 1–3 yr. The GFD patients had become negative for anti-transglutaminase antibodies and recovered a normal or subnormal villous architecture. 16 individuals undergoing either gastric bypass for morbid obesity or endoscopies and biopsies for functional intestinal disorders of nonceliac origin, as described previously (37, 60), were studied as controls. Lymphocytes and epithelial cells were isolated from biopsies or surgical specimens. PBLs were isolated from whole blood from celiac patients and healthy donors. Standard diagnostic tests were used to identify among healthy donors individuals with circulating IgG antibodies against CMV (Abbot Laboratories). All subjects gave written informed consent, and research was approved by institutional review boards.
Cell isolation, cell lines generation, and cell culture.
IE-CTLs and enterocytes were purified from jejunal biopsies as described previously (4). PBLs were isolated from whole blood after Ficoll density gradient centrifugation (GE Healthcare). After sorting, NKG2C+NKG2D+TCR-αβ+CD8+ IE-CTLs and PB-CTLs, and NKG2C−NKG2D+TCR-αβ+CD8+ IE-CTL lines or clones were obtained from celiac patients and one CMV-seropositive individual, as described previously (4). Epithelial intestinal cell lines HUTU-80, INT-407, and HT-29, the NK target cell lines CEM, DAUDI, RAJI, and K562, and the FcγR+ mouse mastocytoma P815 (American Type Culture Collection) were cultured in RPMI-1640 medium with 10% FCS and antibiotics.
Flow cytometric analysis.
Fluorochrome-conjugated anti-CD3, anti-CD8, anti–TCR-αβ, anti-CD103, anti-CD94, anti–IFN-γ, and anti-CD107a were purchased from BD Biosciences. Anti-NKG2A, anti-NKp44, and anti-NKp46 were purchased from Immunotech. Anti-NKG2C (clone 591) was from R&D Systems, and FITC-conjugated anti-epithelial–specific antigen was from Biomeda. For TCR repertoire analysis, PE-conjugated anti-TCRVβ antibodies were purchased from BD Biosciences. Anti-DAP12 (DX37; reference 30) and anti-HLA-E (3D12; reference 61) antibodies as well as HLA-E tetramers loaded with the HLA-G leader peptide (32) were generated.
For surface staining, cells were incubated with fluorochrome-conjugated antibodies according to standard protocols. Unconjugated antibodies were revealed with appropriate fluorochrome-conjugated F (ab′)2 goat anti–mouse IgG isotype from SouthernBiotech.
For intracellular staining, cells were fixed and permeabilized using a BD Biosciences kit. Fluorescence was analyzed on a four-color FACSCalibur (Becton Dickinson) with quadrants set to score as negative >99% of control Ig-stained cells.
Immunohistochemistry.
HLA-E staining was performed as described previously (62), using the anti–HLA-E antibody 3D12 and IgG1 isotype-matched control antibody on 4-μm cryostat sections from small intestinal biopsies embedded in OCT compound (Sakura Fine Technologies) and snap-frozen in liquid nitrogen. Antibody binding was detected using horseradish peroxidase–labeled secondary anti–mouse IgG antibody (EnVision System; DakoCytomation) and DAB chromogen. Sections were counterstained with Gill's 3 hematoxylin, and coverslips were attached by using Sub-X mounting medium (Surgipath).
Cytotoxicity and degranulation assays.
51Chromium-release assays were performed as described previously (4), using intestinal epithelial target cells and the mouse Fcγ+ mastocytoma P815 cell lines at an E/T ratio of 50:1, unless otherwise stated. All experiments were performed in triplicate, and error bars were <10%. In some experiments, epithelial cells or K562 cells were cultured with 15 ng/ml IFN-γ (BD Biosciences) for 12 h. For antibody-dependent cytotoxicity, effectors and targets were incubated in the presence of soluble anti-NKG2C, anti-CD3, or control mouse IgG1 mAbs. Chromium release was measured using a scintillation counter (Packard Instrument Co.). The percentage of specific cytotoxicity was calculated using the following formula: 100 × (cpm experimental − cpm spontaneous)/(cpm maximum − cpm spontaneous).
Degranulation was assayed using flow cytometric quantification of the surface mobilization of CD107a, a membrane protein present in cytolytic granules, as described previously (63, 64). In brief, T cells were stimulated with plate-bound anti-NKp44 (clone 253415; R&D Systems), anti-NKp46 (clone 195314; R&D Systems), anti-CD3 (BD Biosciences) mAbs, or isotype control (BD Biosciences) for 4 h at 37°C. Cells were then stained with anti-CD107a mAb in PBS, 2 mM EDTA.
IFN-γ production.
Freshly isolated cells or NKG2C+ T cell clones were cultured for 12 h with brefeldin A (GolgiPlug; BD Biosciences) in flat-bottom 96-well plates precoated overnight with 10 μg/ml anti-CD3, anti-NKG2D (1D11), anti-NKG2C (591), anti-NKp44 (clone 253415), or anti-NKp46 (clone 195314) antibody and a mouse IgG isotype-matched control, alone or in combination. Cells were fixed and permeabilized using a BD Biosciences kit, stained with FITC-conjugated anti–IFN-γ antibody or isotype-matched control IgG, and analyzed by flow cytometry.
Proliferation assay.
Intraepithelial lymphocyte clones (2 × 105 cells/well) were cultured in 96-well flat-bottomed plates precoated with anti-CD3 or anti-NKG2C antibody at the indicated concentration, alone or in association with anti-NKG2D antibody or control Ig at 10 μg/ml. After 48 h of culture, [3H]thymidine (0.5 mCi/well; MP Biomedicals) was added and plates were cultured for an additional 24 h. The results are shown as the mean of triplicate wells. Error bars were always <10%.
ZAP-70 phosphorylation.
CD8+TCR-αβ+NKG2C+ clones were serum starved 30 h before stimulation. To cross-link immunoreceptors, cells were incubated at 37°C with isotype-matched control Ig, anti-CD3, anti-NKG2D, or anti-NKG2C before adding goat anti–mouse IgG F (ab′)2 fragments for the indicated duration. Cells were lysed for 20 min in a lysis buffer containing freshly added protease and phosphatase inhibitors (50 mM Tris HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and protease inhibitor cocktail tablets). Equal amounts of protein were subjected to SDS-PAGE under reducing conditions in 10% gels, transferred to nitrocellulose membranes (Bio-Rad Laboratories), and probed with anti-phospho–ZAP-70 serum (Cell Signaling Technology) followed by horseradish peroxidase–conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories). To control for loading differences, blots were stripped and reprobed with an anti–ZAP-70 antibody (Cell Signaling Technology). Binding of secondary antibodies was visualized using the enhanced chemiluminescence ECL kit from GE Healthcare.
Microarray hybridization.
Using a standard GeneChip preparation protocol (Affymetrix, Inc.; reference 65), gene array hybridizations and data collection on selected NKG2C+ and NKG2C− IE-CTL and PB-CTL samples were preformed at the Functional Genomics Facility, University of Chicago. In brief, 10 μg total RNA, extracted using the RNeasy Mini kit (QIAGEN), was used to generate double-stranded cDNA using a T7-linked oligo dT primer and the Superscript II RT system (Invitrogen). Purified cDNA was then used to generate biotin-labeled cRNA using the BioArray High Yield RNA Transcript Labeling kit (Enzo Diagnostics) according to the manufacturer's protocol. This biotinylated cRNA was then fragmented and hybridized to Affymetrix GeneChip HG-U133. The arrays were then washed and stained with phycoerythrin-conjugated streptavidin according to the Affymetrix GeneChip protocol and scanned using the Affymetrix Agilent GeneArray Scanner.
Microarray data analysis.
Data analyses were preformed using DNA-Chip Analyzer 1.3 (dChip; reference 65) with the *.CEL files obtained from GeneChip Operating System 1.3 (GCOS 1.3). A PM-only model was used to estimate gene expression level, and the invariant set approach was used for normalization. For comparison analyses of the paired NKG2C+ and NKG2C− populations, thresholds for selecting significant genes were set at a relative difference of >1.5 fold, absolute difference of >100, and a significant paired t test for the signal intensity difference at P < 0.05. Duplicate genes and genes of unknown function were removed. The remaining genes were then grouped based on function, and cluster analysis was preformed using dChip (65).
TCR repertoire analysis.
RNA was isolated and first-strand cDNA template was prepared from CD94+ (control) and NKG2C+ IE-CTLs and PB-CTLs as described previously (36). Using standardized amounts of starting template, TCR-β chain variable region (BV) PCR was performed as described previously (49). For nucleotide sequence analysis of certain TCR BV family repertoires, selected BV region families were cloned and sequenced, as described previously (49). The 295 TCR-β chain nucleotide sequences obtained were analyzed within each BV family, and CDR3 motifs with identical sequences were grouped together to show TCR clonotypes as described previously (49). For simplicity, a clone was considered expanded if two or more identical TCR-β chain sequences were identified during sequence analysis (49). In this study, we used the TCR BV nomenclature proposed by Arden et al. (66) for correlation with the anti-BV mAb specificities. Correspondence between the Arden and HGNC nomenclatures can be found at http://imgt.cines.fr (67).
Quantitative RT-PCR.
Quantitative real-time RT-PCR was preformed using the iCycler iQ real-time PCR detection system (Bio-Rad Laboratories) and an SYBR green amplification kit (PE Biosystems). To confirm that the same amount of starting template was used in each BV PCR reaction, the quantity was standardized using GAPDH primers, as described previously (4). For comparison of the relative levels of TCR expression between NKG2C+ and control CD94+ IE-CTL populations, we used primers specific for the constant region of the TCR-β chain and normalized their expression against GAPDH levels, as described previously (4), using 10 μm of the following primers with an annealing temperature of 56°C: 5′-AGCCCGCCCTCAATGACTCCAGATACT-3′ and 5′-ATA-GAGGATGGTGGCAGACAGGAC-3′.
Statistical analysis.
All expression data were transformed from percentages to natural logarithms of the percentages to reduce the heterogeneity of variance across the AC disease, GFD, and control groups. Analyses of variance 2-degree-of-freedom F-tests were constructed to test for differences in log expression across the three groups. Where F-tests were significant at the α = 0.05 level, mean pairwise comparisons were tested with a Tukey multiple comparisons correction.
A Fisher's exact test was used to determine the probability that 19 out of 46 genes found to be up-regulated by microarray analysis were NK- related genes. This test conditions on the fact that 16,500 were functional genes, of which 150 were NK genes, and that 46 genes of the 16,500 were up-regulated in NKG2C+ CTLs. The p-value reports the probability that 19 or more NK genes would be significantly expressed under the null hypothesis that NK genes are not overexpressed in NKG2C+ cells as compared with NKG2C− cells. All statistic analysis was performed using SAS (SAS 2002–2005; SAS Institute).
Online supplemental material.
The sequence of all TCR-β chain clonotypes obtained in this study can be accessed with the following accession numbers: DQ473356 and DQ473390. All microarray data can be accessed using the National Center for Biotechnology Information GEO accession number GSE4592. Table S1 shows data expressed as fold-change for selected genes that were obtained when comparing separately IE-CTL and PB-CTL NKG2C+ and NKG2C− celiac clones. Table S1 is available at http://www.jem.org/cgi/content/full/jem.20060028/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank Albert Bendelac for critical reading of the manuscript, Hatice Aldemir for preparation of HLA-E tetramers, Theodore Karrison for help with statistical analysis, Xinmin Li for microarray analysis, and Ryan Duggan for cell sorting. We would also like to thank the University of Chicago Celiac Disease Center, The Columbia Presbyterian Hospital Celiac Disease Center, and the University of Chicago DDRC for support.
This work was supported by RO1 DK58727, RO1 DK063158, and R01 CA89294 (to L.L. Lanier). L.L. Lanier is an American Cancer Society Research Professor.
The authors have no conflicting financial interests.
Abbreviations used: AC, active celiac; EATL, enteropathy-associated T cell lymphoma; GFD, gluten-free diet; IE-CTL, intraepithelial CTL; ITAM, immunoreceptor tyrosine-based activation motif; PB-CTL, peripheral blood CTL.
References
- 1.Jabri, B., and B. Meresse. 2006. NKG2 receptor-mediated regulation of effector CTL functions in the human tissue microenvironment. Curr. Top. Microbiol. Immunol. 298:139–156. [DOI] [PubMed] [Google Scholar]
- 2.Roberts, A.I., L. Lee, E. Schwarz, V. Groh, T. Spies, E.C. Ebert, and B. Jabri. 2001. NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 167:5527–5530. [DOI] [PubMed] [Google Scholar]
- 3.Ogasawara, K., J.A. Hamerman, L.R. Ehrlich, H. Bour-Jordan, P. Santamaria, J.A. Bluestone, and L.L. Lanier. 2004. NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity. 20:757–767. [DOI] [PubMed] [Google Scholar]
- 4.Meresse, B., Z. Chen, C. Ciszewski, M. Tretiakova, G. Bhagat, T.N. Krausz, D.H. Raulet, L.L. Lanier, V. Groh, T. Spies, et al. 2004. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 21:357–366. [DOI] [PubMed] [Google Scholar]
- 5.Hue, S., J.J. Mention, R.C. Monteiro, S. Zhang, C. Cellier, J. Schmitz, V. Verkarre, N. Fodil, S. Bahram, N. Cerf-Bensussan, and S. Caillat-Zucman. 2004. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity. 21:367–377. [DOI] [PubMed] [Google Scholar]
- 6.Groh, V., A. Bruhl, H. El-Gabalawy, J.L. Nelson, and T. Spies. 2003. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA. 100:9452–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, S., V. Groh, J. Wu, A. Steinle, J.H. Phillips, L.L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 285:727–729. [DOI] [PubMed] [Google Scholar]
- 8.Cosman, D., J. Mullberg, C.L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N.J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 14:123–133. [DOI] [PubMed] [Google Scholar]
- 9.Carayannopoulos, L.N., O.V. Naidenko, D.H. Fremont, and W.M. Yokoyama. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169:4079–4083. [DOI] [PubMed] [Google Scholar]
- 10.Cerwenka, A., A.B. Bakker, T. McClanahan, J. Wagner, J. Wu, J.H. Phillips, and L.L. Lanier. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727. [DOI] [PubMed] [Google Scholar]
- 11.Diefenbach, A., J.K. Hsia, M.Y. Hsiung, and D.H. Raulet. 2003. A novel ligand for the NKG2D receptor activates NK cells and macrophages and induces tumor immunity. Eur. J. Immunol. 33:381–391. [DOI] [PubMed] [Google Scholar]
- 12.Wu, J., Y. Song, A.B. Bakker, S. Bauer, T. Spies, L.L. Lanier, and J.H. Phillips. 1999. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 285:730–732. [DOI] [PubMed] [Google Scholar]
- 13.Rosen, D.B., M. Araki, J.A. Hamerman, T. Chen, T. Yamamura, and L.L. Lanier. 2004. A structural basis for the association of DAP12 with mouse, but not human, NKG2D. J. Immunol. 173:2470–2478. [DOI] [PubMed] [Google Scholar]
- 14.Andre, P., R. Castriconi, M. Espeli, N. Anfossi, T. Juarez, S. Hue, H. Conway, F. Romagne, A. Dondero, M. Nanni, et al. 2004. Comparative analysis of human NK cell activation induced by NKG2D and natural cytotoxicity receptors. Eur. J. Immunol. 34:961–971. [DOI] [PubMed] [Google Scholar]
- 15.Diefenbach, A., E. Tomasello, M. Lucas, A.M. Jamieson, J.K. Hsia, E. Vivier, and D.H. Raulet. 2002. Selective associations with signaling proteins determine stimulatory versus costimulatory activity of NKG2D. Nat. Immunol. 3:1142–1149. [DOI] [PubMed] [Google Scholar]
- 16.Gilfillan, S., E.L. Ho, M. Cella, W.M. Yokoyama, and M. Colonna. 2002. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat. Immunol. 3:1150–1155. [DOI] [PubMed] [Google Scholar]
- 17.Fehniger, T.A., and M.A. Caligiuri. 2001. Interleukin 15: biology and relevance to human disease. Blood. 97:14–32. [DOI] [PubMed] [Google Scholar]
- 18.Sollid, L.M. 2002. Coeliac disease: dissecting a complex inflammatory disorder. Nat. Rev. Immunol. 2:647–655. [DOI] [PubMed] [Google Scholar]
- 19.Koning, F. 2005. Celiac disease: caught between a rock and a hard place. Gastroenterology. 129:1294–1301. [DOI] [PubMed] [Google Scholar]
- 20.Marietta, E., K. Black, M. Camilleri, P. Krause, R.S. Rogers III, C. David, M.R. Pittelkow, and J.A. Murray. 2004. A new model for dermatitis herpetiformis that uses HLA-DQ8 transgenic NOD mice. J. Clin. Invest. 114:1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jabri, B., D.D. Kasarda, and P.H. Green. 2005. Innate and adaptive immunity: the yin and yang of celiac disease. Immunol. Rev. 206:219–231. [DOI] [PubMed] [Google Scholar]
- 22.Green, P.H., and B. Jabri. 2003. Coeliac disease. Lancet. 362:383–391. [DOI] [PubMed] [Google Scholar]
- 23.Maiuri, L., C. Ciacci, I. Ricciardelli, L. Vacca, V. Raia, A. Rispo, M. Griffin, T. Issekutz, S. Quaratino, and M. Londei. 2005. Unexpected role of surface transglutaminase type II in celiac disease. Gastroenterology. 129:1400–1413. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S.K., W. Lo, L. Memeo, H. Rotterdam, and P.H. Green. 2003. Duodenal histology in patients with celiac disease after treatment with a gluten-free diet. Gastrointest. Endosc. 57:187–191. [DOI] [PubMed] [Google Scholar]
- 25.Wahab, P.J., J.W. Meijer, and C.J. Mulder. 2002. Histologic follow-up of people with celiac disease on a gluten-free diet: slow and incomplete recovery. Am. J. Clin. Pathol. 118:459–463. [DOI] [PubMed] [Google Scholar]
- 26.Forsberg, G., O. Hernell, S. Melgar, A. Israelsson, S. Hammarstrom, and M.L. Hammarstrom. 2002. Paradoxical coexpression of proinflammatory and down-regulatory cytokines in intestinal T cells in childhood celiac disease. Gastroenterology. 123:667–678. [DOI] [PubMed] [Google Scholar]
- 27.Olaussen, R.W., F.E. Johansen, K.E. Lundin, J. Jahnsen, P. Brandtzaeg, and I.N. Farstad. 2002. Interferon-gamma-secreting T cells localize to the epithelium in coeliac disease. Scand. J. Immunol. 56:652–664. [DOI] [PubMed] [Google Scholar]
- 28.Isaacson, P.G. 2000. Relation between cryptic intestinal lymphoma and refractory sprue. Lancet. 356:178–179. [DOI] [PubMed] [Google Scholar]
- 29.Daum, S., C. Cellier, and C.J. Mulder. 2005. Refractory coeliac disease. Best Pract. Res. Clin. Gastroenterol. 19:413–424. [DOI] [PubMed] [Google Scholar]
- 30.Lanier, L.L., B. Corliss, J. Wu, and J.H. Phillips. 1998. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 8:693–701. [DOI] [PubMed] [Google Scholar]
- 31.Tomasello, E., and E. Vivier. 2005. KARAP/DAP12/TYROBP: three names and a multiplicity of biological functions. Eur. J. Immunol. 35:1670–1677. [DOI] [PubMed] [Google Scholar]
- 32.Braud, V.M., D.S. Allan, C.A. O'Callaghan, K. Soderstrom, A. D'Andrea, G.S. Ogg, S. Lazetic, N.T. Young, J.I. Bell, J.H. Phillips, et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 391:795–799. [DOI] [PubMed] [Google Scholar]
- 33.Lee, N., M. Llano, M. Carretero, A. Ishitani, F. Navarro, M. Lopez-Botet, and D.E. Geraghty. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA. 95:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guma, M., L.K. Busch, L.I. Salazar-Fontana, B. Bellosillo, C. Morte, P. Garcia, and M. Lopez-Botet. 2005. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur. J. Immunol. 35:2071–2080. [DOI] [PubMed] [Google Scholar]
- 35.Guma, M., A. Angulo, C. Vilches, N. Gomez-Lozano, N. Malats, and M. Lopez-Botet. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 104:3664–3671. [DOI] [PubMed] [Google Scholar]
- 36.Jabri, B., J.M. Selby, H. Negulescu, L. Lee, A.I. Roberts, A. Beavis, M. Lopez-Botet, E.C. Ebert, and R.J. Winchester. 2002. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity. 17:487–499. [DOI] [PubMed] [Google Scholar]
- 37.Jabri, B., N.P. de Serre, C. Cellier, K. Evans, C. Gache, C. Carvalho, J.F. Mougenot, M. Allez, R. Jian, P. Desreumaux, et al. 2000. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 118:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arlettaz, L., J. Villard, C. de Rham, S. Degermann, B. Chapuis, B. Huard, and E. Roosnek. 2004. Activating CD94:NKG2C and inhibitory CD94:NKG2A receptors are expressed by distinct subsets of committed CD8+ TCR alphabeta lymphocytes. Eur. J. Immunol. 34:3456–3464. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Botet, M., J.J. Perez-Villar, M. Carretero, A. Rodriguez, I. Melero, T. Bellon, M. Llano, and F. Navarro. 1997. Structure and function of the CD94 C-type lectin receptor complex involved in recognition of HLA class I molecules. Immunol. Rev. 155:165–174. [DOI] [PubMed] [Google Scholar]
- 40.Kaiser, B.K., F. Barahmand-Pour, W. Paulsene, S. Medley, D.E. Geraghty, and R.K. Strong. 2005. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 174:2878–2884. [DOI] [PubMed] [Google Scholar]
- 41.Brumbaugh, K.M., J.J. Perez-Villar, C.J. Dick, R.A. Schoon, M. Lopez-Botet, and P.J. Leibson. 1996. Clonotypic differences in signaling from CD94 (kp43) on NK cells lead to divergent cellular responses. J. Immunol. 157:2804–2812. [PubMed] [Google Scholar]
- 42.Rinke de Wit, T.F., S. Vloemans, P.J. van den Elsen, A. Haworth, and P.L. Stern. 1990. Differential expression of the HLA class I multigene family by human embryonal carcinoma and choriocarcinoma cell lines. J. Immunol. 144:1080–1087. [PubMed] [Google Scholar]
- 43.Cellier, C., N. Patey, L. Mauvieux, B. Jabri, E. Delabesse, J.P. Cervoni, M.L. Burtin, D. Guy-Grand, Y. Bouhnik, R. Modigliani, et al. 1998. Abnormal intestinal intraepithelial lymphocytes in refractory sprue. Gastroenterology. 114:471–481. [DOI] [PubMed] [Google Scholar]
- 44.Vitale, M., C. Bottino, S. Sivori, L. Sanseverino, R. Castriconi, E. Marcenaro, R. Augugliaro, L. Moretta, and A. Moretta. 1998. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex–restricted tumor cell lysis. J. Exp. Med. 187:2065–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanier, L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359–393. [DOI] [PubMed] [Google Scholar]
- 46.Raulet, D.H., R.E. Vance, and C.W. McMahon. 2001. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 19:291–330. [DOI] [PubMed] [Google Scholar]
- 47.Yokoyama, W.M., and B.F. Plougastel. 2003. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 3:304–316. [DOI] [PubMed] [Google Scholar]
- 48.Chiesa, S., E. Tomasello, E. Vivier, and F. Vely. 2005. Coordination of activating and inhibitory signals in natural killer cells. Mol. Immunol. 42:477–484. [DOI] [PubMed] [Google Scholar]
- 49.Curran, S.A., O.M. FitzGerald, P.J. Costello, J.M. Selby, D.J. Kane, B. Bresnihan, and R. Winchester. 2004. Nucleotide sequencing of psoriatic arthritis tissue before and during methotrexate administration reveals a complex inflammatory T cell infiltrate with very few clones exhibiting features that suggest they drive the inflammatory process by recognizing autoantigens. J. Immunol. 172:1935–1944. [DOI] [PubMed] [Google Scholar]
- 50.Biassoni, R., C. Cantoni, D. Pende, S. Sivori, S. Parolini, M. Vitale, C. Bottino, and A. Moretta. 2001. Human natural killer cell receptors and co-receptors. Immunol. Rev. 181:203–214. [DOI] [PubMed] [Google Scholar]
- 51.Lanier, L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. [DOI] [PubMed] [Google Scholar]
- 52.Brooks, C.G. 1983. Reversible induction of natural killer cell activity in cloned murine cytotoxic T lymphocytes. Nature. 305:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks, C.G., M. Holscher, and D. Urdal. 1985. Natural killer activity in cloned cytotoxic T lymphocytes: regulation by interleukin 2, interferon, and specific antigen. J. Immunol. 135:1145–1152. [PubMed] [Google Scholar]
- 54.Shustik, C., I.R. Cohen, R.S. Schwartz, E. Latham-Griffin, and S.D. Waksal. 1976. T lymphocytes with promiscuous cytotoxicity. Nature. 263:699–701. [DOI] [PubMed] [Google Scholar]
- 55.Farstad, I.N., F.E. Johansen, L. Vlatkovic, J. Jahnsen, H. Scott, O. Fausa, A. Bjorneklett, P. Brandtzaeg, and T.S. Halstensen. 2002. Heterogeneity of intraepithelial lymphocytes in refractory sprue: potential implications of CD30 expression. Gut. 51:372–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mention, J.J., M. Ben Ahmed, B. Begue, U. Barbe, V. Verkarre, V. Asnafi, J.F. Colombel, P.H. Cugnenc, F.M. Ruemmele, E. McIntyre, et al. 2003. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology. 125:730–745. [DOI] [PubMed] [Google Scholar]
- 57.Karre, K. 2002. NK cells, MHC class I molecules and the missing self. Scand. J. Immunol. 55:221–228. [DOI] [PubMed] [Google Scholar]
- 58.McMahon, C.W., and D.H. Raulet. 2001. Expression and function of NK cell receptors in CD8+ T cells. Curr. Opin. Immunol. 13:465–470. [DOI] [PubMed] [Google Scholar]
- 59.Ortega, C., P. Romero, A. Palma, T. Orta, J. Pena, A. Garcia-Vinuesa, I.J. Molina, and M. Santamaria. 2004. Role for NKG2-A and NKG2-C surface receptors in chronic CD4+ T-cell responses. Immunol. Cell Biol. 82:587–595. [DOI] [PubMed] [Google Scholar]
- 60.Taunk, J., A.I. Roberts, and E.C. Ebert. 1992. Spontaneous cytotoxicity of human intraepithelial lymphocytes against epithelial cell tumors. Gastroenterology. 102:69–75. [DOI] [PubMed] [Google Scholar]
- 61.Lee, N., D.R. Goodlett, A. Ishitani, H. Marquardt, and D.E. Geraghty. 1998. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160:4951–4960. [PubMed] [Google Scholar]
- 62.Ishitani, A., N. Sageshima, N. Lee, N. Dorofeeva, K. Hatake, H. Marquardt, and D.E. Geraghty. 2003. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J. Immunol. 171:1376–1384. [DOI] [PubMed] [Google Scholar]
- 63.Rubio, V., T.B. Stuge, N. Singh, M.R. Betts, J.S. Weber, M. Roederer, and P.P. Lee. 2003. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat. Med. 9:1377–1382. [DOI] [PubMed] [Google Scholar]
- 64.Bryceson, Y.T., M.E. March, D.F. Barber, H.G. Ljunggren, and E.O. Long. 2005. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 202:1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li, X., R.J. Quigg, J. Zhou, J.T. Ryaby, and H. Wang. 2005. Early signals for fracture healing. J. Cell. Biochem. 95:189–205. [DOI] [PubMed] [Google Scholar]
- 66.Arden, B., S.P. Clark, D. Kabelitz, and T.W. Mak. 1995. Human T-cell receptor variable gene segment families. Immunogenetics. 42:455–500. [DOI] [PubMed] [Google Scholar]
- 67.Lefranc, M.P., V. Giudicelli, Q. Kaas, E. Duprat, J. Jabado-Michaloud, D. Scaviner, C. Ginestoux, O. Clement, D. Chaume, and G. Lefranc. 2005. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 33:D593–D597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.