Abstract
The crystal structure of the tetrameric DNA-binding domain of the single-stranded DNA binding protein from Escherichia coli was determined at a resolution of 2.9 Å using multiwavelength anomalous dispersion. Each monomer in the tetramer is topologically similar to an oligomer-binding fold. Two monomers each contribute three β-strands to a single six-stranded β-sheet to form a dimer. Two dimer–dimer interfaces are observed within the crystal. One of these stabilizes the tetramer in solution. The other interface promotes a superhelical structure within the crystal that may reflect tetramer–tetramer interactions involved in the positive cooperative binding of the single-stranded DNA-binding protein to single-stranded DNA.
Keywords: x-ray crystal structure/DNA replication/helix destabilizing
Single-stranded DNA-binding (SSB) proteins are a class of nonspecific DNA binding proteins that play essential roles in DNA metabolism. Most organisms, including many bacteriophage and viruses, encode their own SSB proteins, all of which bind preferentially and with high affinity to single-stranded DNA (ssDNA).
The Escherichia coli SSB protein (1) and the bacteriophage T4 gene 32 protein (T4 gp32) (2) were the first SSBs identified and remain the best studied examples of this class of protein (3–5). Although these two proteins share functional similarities, they display significant differences in their ssDNA binding properties, due at least in part, to the differences in their quaternary structure (6). T4 gp32 is a stable monomer (2), whereas the E. coli SSB protein is a stable homotetramer and the tetramer is the functional form of the protein (7).
E. coli SSB displays multiple modes of binding to single-stranded polynucleotides, referred to as (SSB)n, that differ in the number of nucleotides (n) occluded by each bound tetramer (5). At least three binding modes have been identified with n = 35 ± 2, n = 56 ± 3, and n = 65 ± 3 nucleotides per tetramer (8–10). Only two of the subunits of the tetramer interact with ssDNA in the (SSB)35 mode, whereas all four subunits interact with ssDNA in the (SSB)65 mode (11, 12). In at least one of these modes [(SSB)65], the ssDNA wraps around the SSB tetramer (8, 13, 14).
A common feature of SSB proteins is their ability to bind with positive cooperativity to single-stranded polynucleotides and thus form clusters of protein, even at low binding densities. However, the type and magnitude of the positive cooperativity observed for E. coli SSB differs dramatically for the different binding modes. SSB tetramers bind with an “unlimited” type of inter-tetramer cooperativity in the (SSB)35 mode, and thus can form long protein clusters which can saturate the DNA (10, 15). In contrast, binding in the (SSB)65 mode occurs with a “limited” type of positive inter-tetramer cooperativity, such that “beaded” structures corresponding to tetramers and dimers of tetramers (“octamers”) are observed by electron microscopy (14–16). These different modes may be used selectively in replication, recombination, and repair (6).
Each SSB monomer consists of 177 amino acids (Mr = 18,843) (17). Proteolysis studies of the intact SSB tetramer indicate that the ssDNA binding site is contained within the first 115 N-terminal amino acids (18). N-terminal fragments resulting from cleavage by trypsin (SSBT) after Arg-115 or by chymotrypsin (SSBC) after Trp-135 also form tetramers and bind ssDNA. Interestingly, mitochondrial SSB proteins share significant similarities to E. coli SSB within the first 100 N-terminal residues (19). Recently, the structure of the homotetrameric human mitochondrial SSB (hmtSSB) has been determined and is expected to be very similar to that of the DNA binding domain of E. coli SSB (20).
E. coli SSB has been the subject of extensive crystallization efforts (21–25). The first reports described the crystallization of full length E. coli SSB (21, 22). The asymmetric unit in these crystals contained two native and two proteolytically degraded subunits with D2 symmetry. Later reports described other crystal forms of the full-length protein as well as the chymotryptic and tryptic fragments of E. coli SSB. However, a structure remained to be determined. We report here the crystal structure of the DNA binding domain of the E. coli SSB tetramer corresponding to its chymotryptic fragment (SSBC).
MATERIALS AND METHODS
Selenomethionine-containing chymotryptic fragment of E. coli SSB was obtained using previously described methods (18, 26, 27). Crystals were grown in a hanging drop against a reservoir of 0.1 M Tris⋅HCl (pH 8.5) and 5% (wt/vol) PEG 1000. After cryoprotection and freezing to liquid nitrogen temperature, these crystals diffracted to 2.7-Å resolution at the National Synchrotron Light Source (Brookhaven National Laboratory). Crystals were in space group C2, with unit cell dimensions a = 58.0 Å, b = 105.8 Å, c = 90.2 Å, and β = 99.0°. The self-rotation function indicated the presence of a noncrystallographic 2-fold axis. A native data set was collected at the Stanford Synchrotron Radiation Laboratory at room temperature from a single crystal to a resolution of 2.7 Å (Table 1).
Table 1.
Data collection
| Wavelength, Å | Reflection, N | Redundancy | Completeness, %, (I > 1σ) | (〈I/σ(I)〉) | Rsym,* % |
|---|---|---|---|---|---|
| MAD data collection statistics (30.0 2.7 Å) | |||||
| 0.9879 | 14,346 | 5.4 | 95.9 | 21.3 | 6.6 |
| 0.9793 | 14,262 | 5.4 | 95.8 | 20.6 | 7.2 |
| 0.9792 | 14,268 | 5.3 | 95.4 | 20.4 | 7.9 |
| 0.9686 | 14,215 | 5.3 | 95.4 | 20.1 | 7.4 |
| Native data collection statistics (30.0 2.7 Å) | |||||
| 1.0800 | 12,833 | 2.6 | 83.6 | 12.7 | 5.3 |
| Refinement and stereochemical statistics (8–2.9 Å)† | |||||
| R-factor, % 23.0 | |||||
| Free R-factor (%) 29.5‡ | |||||
| Reflections, |F| > 1σ 10,473 (89.7%, 75.6%)§ | |||||
| Total number of atoms 3,044 | |||||
| rms deviation in bond length, Å 0.017 | |||||
| rms deviation in bond angles, ° 2.0 | |||||
Rsym = ∑|I − 〈I〉|/∑ I where I = observed intensity and 〈I〉 = average intensity from multiple observations of symmetry related reflections.
Refinement was carried out using noncrystallographic restraints of 200 kcal/mol per Å2 on all four monomers and all residues but those of the L45 loop (85–99), where restraints of 50 kcal/mol per A2 were applied.
Free R-values were calculated using 10% of the data set (|F| > 1σ).
Percentage of overall completeness and completeness in the last resolution shell (2.9–3.0 Å), respectively. Data beyond 2.9 Å were not used for refinement because of lack of completeness.
Multiwavelength anomalous diffraction (MAD) data were collected at National Synchrotron Light Source (Table 1) from a single selenomethionine-substituted crystal. All diffraction images were processed using the program denzo and scaled with the program scalepack (28). Data were used to obtain a bayesian estimate of the magnitudes of partial structure factors corresponding to the anomalously scattering atoms, FA [program madbest (29, 30)]. Values for f′ and f" for selenium atoms were obtained from Hall et al. (31). The resulting values of FA were used in a difference Patterson synthesis [program hassp (32)] that yielded the positions of six selenium atoms. The positions and occupancies of the six selenium atoms were refined against a pseudo-SIRAS (single iomorphous replacement with anomalous scattering) data, as described and implemented in the program heavy version 4 (33). Two selenium atoms refined to low occupancy and therefore only four selenium atoms were used in the phase calculation (33). Two of these atoms were related to the two remaining heavy metals by a noncrystallographic 2-fold axis with rotational angle values similar to those found in the self-rotation function analysis. Calculation of phases from 15 to 3.0 Å indicated a rapid fall-off of the figure of merit beyond 4 Å. Therefore, a protocol of phase calculation to a resolution of 4 Å followed by noncrystallographic symmetry averaging combined with solvent flattening and phase extension to 3.1 Å was implemented [program dm (34)].
A partial model using a poly(A) chain was built using the program o and a database of protein structures (35). During this process, Cα atom positions of partial atomic models were repeatedly used to improve the phases by phase combination using sigmaa (36) and solvent flattening. Refinement [program xplor (37, 38)] proceeded using the native data set (Table 1). The four selenium positions used in phasing could be easily interpretated as corresponding to Met-109 in all four monomers. However, an ambiguity remained in the interpretation of the two weak selenium positions. Sequence alignment of hmtSSB and E. coli SSB and reference to the hmtSSB structure (20) suggested that these positions corresponded to Met-23 and a model was built using these methionine positions as “anchors” for the building of side chains. Subsequent least-squares refinement and simulated annealing refinement (39) with all four monomers restrained by noncrystallographic symmetry (see Table 1) and restrained refinement of temperature factors (see below) resulted in a model with a free R-value of 29.5%, and an R-value of 23.0%. rms deviation in main chain atoms between monomers is between 0.04 and 0.06 Å. Average temperature factor is 23 Å2 with an rms deviation for temperature factors of bonded atoms of 1.65 Å2. The present model contains 96 residues out of 135 residues from residues 3 to 23, 28 to 39, and 50 to 112 and does not include any solvent molecules. It has unbroken backbone electron density in the regions traced (Fig. 1). All residues are in the allowed regions of the Ramachandran plot (40).
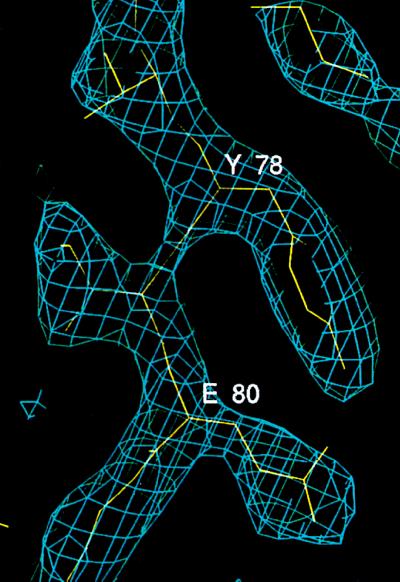
Figure 1.
Electron density for a representative region of the SSBC structure. The protein model corresponding to strand 4 is shown in yellow. Light blue contour lines indicate electron density at 1.2 σ above the mean density, in a map calculated using coefficients (2|Fo| − |Fc|)exp(−iαc), where |Fo| is the observed structure factor amplitude, and |Fc| and αc are the amplitudes and phases calculated from the model.
RESULTS AND DISCUSSION
Structures of the SSBC Monomer and Dimer.
The SSBC monomer is topologically identical to a number of proteins known to bind oligonucleotides or oligosaccharides and its fold belongs to the well-characterized OB (oligomer-binding) fold (Fig. 2, A and B) (41). For consistency, the participating secondary structural elements of the SSBC monomer are labeled according to Murzin (41) and the intervening loop regions are labeled as Lxy where x and y indicate the numbers of the strands that they join. The core of the monomer structure is hydrophobic in nature. The L23 and L45 loops of the SSBC monomer are very extended and form β-hairpins. Density for the tip of L23 was poorly defined, and therefore this region of the structure was not included in the model.
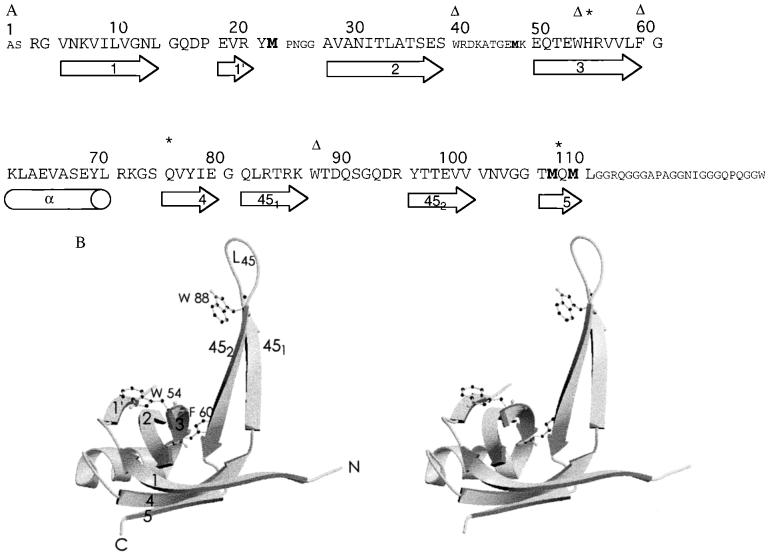
Figure 2.
Primary, secondary, tertiary, and quaternary structures of SSBC. (A) Amino acid sequence of SSBC. Larger letters indicate the region of the crystal structure with well-defined electron density. Amino acid numbers are indicated at the top while the secondary structural elements that they form are shown schematically at the bottom. Notations of secondary structures are as defined by Murzin (41) with the strands of the OB fold labeled 1–5. (Δ), Residues known to be important in binding ssDNA. ∗, Residues that have been identified as involved in the stability of the tetrameric structure of E. coli SSB. Methionine residues are highlighted in bold. In the monomer, strand 1 starts at residue 7 and ends at residue 14. However, residues 5–11 are involved in the dimer interface to form a continuous β-sheet with strand 1 of another monomer. (B) Stereo diagram of the SSBC monomer (42). This diagram illustrates the OB fold and the notation used to describe it. Residues important for ssDNA binding are indicated. (C) Schematic diagram of the SSBC dimer and role of His-55 (42). The unprimed and primed labels identify the secondary structural elements of the first and second monomer, respectively. This stereo view illustrates the monomer–monomer interface as a continuous six-stranded β-sheet. His-55 is shown in ball-and-stick representation. (D) Stereo diagram of the L45-mediated tetramer (52). The labels ′, ", and ‴ identify the second, third, and fourth monomers, respectively, while the absence of these labels identifies the first monomer. (E) Stereo diagram of the six-stranded β-sheet-mediated tetramer (42). This schematic diagram also shows the location of Gln-76 and Gln-110, which when mutated independently to Leu suppress the effect of a His-55 to Lys mutation.
In the SSBC dimer, two antiparallel β-sheets from two SSB subunits form an extended six-stranded antiparallel β-sheet (Fig. 2C). The participating strands are 1, 4, and 5 in each subunit. The monomer–monomer interface therefore consists primarily of mainchain hydrogen bonds involving residues 5–11 of strand 1 in both monomers. In addition, the interface contains contributions from strand 3 [residues His-55 (Fig. 2C) and Glu-53] and strand 1 (Asn-6), and the base of loop L45 (Leu-83 and Thr-99). The total surface area buried upon dimer formation is 1,347 Å2, an area typical of those surveyed for protein–protein interactions (43). The two monomers in the dimer are related by a 2-fold axis.
Although only a few natural mutants of E. coli SSB have been isolated and characterized, one of them, namely ssb-1, which substitutes His-55 with Tyr, maps at the monomer–monomer interface (Fig. 2C). The temperature-sensitive ssb-1 mutation (3) results in a destabilization of the tetramer with respect to monomers (7, 44). Our structural data indicate that His-55 of one monomer is involved in hydrogen bonding contacts with the side chain of Asn-6 and the main chain carbonyl oxygen of Leu-83 within the other monomer in the dimer. Destabilization of the monomer–monomer interface by substitution with the bulkier tyrosine side chain could result from increased steric hindrance and/or changes in the nature of the contact surfaces.
Architecture of the SSBC Tetramer.
SSBC assembles to form tetramers in solution (7). However, two different interfaces between dimers are apparent within the crystal. Fig. 2D shows a tetramer which consists of two dimers sharing a noncrystallographic interface formed primarily by two L45 loops, while Fig. 2E shows another tetramer consisting of two dimers sharing a crystallographic interface formed by two six-stranded β-sheets. We refer to the former configuration as the L45-mediated tetramer, while the latter configuration is referred to as the six-stranded β-sheet-mediated tetramer.
If the L45 loop is seen as a tail, then the two dimers in the L45-mediated tetramer come together in an offset tail-to-tail arrangement. As a consequence, only one L45 loop per dimer is involved in forming the dimer–dimer interface. Indeed, the interface involves the formation of a short antiparallel four-stranded β-sheet consisting of two L45 loops contributed by only one monomer from each dimer (labeled 1 and 1‴ in Fig. 2D). Residues involved are 86–89 from one strand (strand 451 of Fig. 2A) of the β-hairpins that form loop L45. Additional contacts are provided by the tip of each of the L45 loops (residues 91–94) with hydrogen bonding interactions observed between Arg21 and Gln91. The L45-mediated dimer–dimer interface extends over a surface area of about 670 Å2. This tetramer configuration is that seen in the asymmetric unit of the crystal.
In the six-stranded β-sheet-mediated tetramer, the amount of contact surface area between dimers (1,003 Å2) is quantitatively greater than that observed for the L45-mediated tetramer configuration described above. Also, the structural make-up of this interface is different. Contacts involve residues’ side chains rather than mainchain atoms. The interface is mixed in its chemical composition, with a very limited hydrophobic core surrounded by a network of hydrogen-bonded tyrosine, glutamic acid, lysine, and glutamine residues (Lys-7, Tyr-78, Glu-80, Gln-110, Gln-76). Lys-7 and Glu-80 form a salt bridge, a feature also observed in the hmtSSB structure (20). By virtue of the 2-fold axis relating monomers in the dimer and the crystallographic relationship between dimers in the tetramer, the six-stranded β-sheet-mediated tetramer displays D2 symmetry. This tetramer configuration has also been reported for hmtSSB (20).
Which of the two possible tetramer configurations observed in the crystal reflects the configuration of the SSBC tetramer in solution? Studies by Carlini and Porter (45) suggest that the six-stranded β-sheet-mediated tetramer represents the configuration of SSBC in solution. These authors genetically selected for intragenic second site suppressors of a His-55 → Lys mutation that destabilizes the SSB tetramer. The only second site mutations obtained were Gln-110 → Leu and Gln-76 → Leu (45), both of which map to the six-stranded β-sheet-mediated dimer–dimer interface, rather than at the monomer – monomer interface of the dimer, the site of the original mutation (Fig. 2E). This confirms the six-stranded β-sheet as an important interface for tetramerization of SSBC.
Residues and Surfaces Involved in Binding of ssDNA and Implications for Cooperative Binding to ssDNA.
Mutational studies of tryptophans and chemical modification of lysines have indicated that these residues are important for ssDNA binding (46, 47). The role of lysines is supported by the salt-dependence of the binding constant, reflecting substantial electrostatic interactions (48).
There are four tryptophan residues per E. coli SSB subunit at positions 40, 54, 88, and 135; however, only three of them, Trp-40, -54, and -88 appear to play a role in ssDNA binding. Trp-40 and Trp-54 are required for high affinity binding to ssDNA and both of these residues also form stacking interactions with the bases (46, 49). Although mutations at Trp-88 influence DNA binding, this residue does not form such stacking interactions and thus is in a different class than either Trp-40 or Trp-54 (50). Interestingly, mutations at Trp-54 and Trp-88 have a dramatic effect on the relative stabilities of the different SSB–ssDNA binding modes (46). Phe-60 can be photochemically crosslinked to a ssDNA octamer [(dT)8] (51) and mutations at Phe60 also influence ssDNA binding affinity (50).
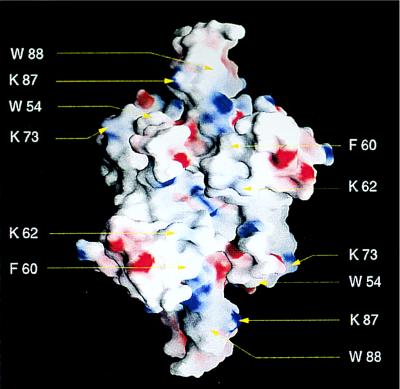
The locations of Trp-54, Trp-88, and Phe-60, as well as all lysine residues, are depicted in Figs. 2B and 3. The amino acids known to be important for binding ssDNA—i.e., Trp-54, and Phe-60—are located on an exposed surface running parallel to strand 3, whereas Trp-88 is located in loop L45. Trp-40 is in the part of the L23 loop which is disordered. The exposed surface between Trp-54 and Phe-60 in each monomer does not contain positively charged patches and therefore is not expected to bind ssDNA. In fact, an understanding as to where the DNA may bind requires inspection of the surface of the tetramer rather than that of the monomer alone. Fig. 3 shows a rendering of the electrostatic potential at the surface of the tetramer in the same orientation as in Fig. 2E. In this orientation, there are surfaces of positive electrostatic potential that may form a path for ssDNA binding that could span the entire structure from side to side (upper left Trp-54 to lower right Trp-54 in Fig. 3). Hence, two such surfaces would be available for binding of ssDNA within the tetramer, that presented in Fig. 3 and the equivalent surface on the side opposite.
Figure 3.
Surfaces involved in binding of ssDNA to SSBC. The molecular surface of the the tetramer in the orientation of Fig. 2E, calculated and displayed using grasp (52). The surface is colored deep blue (15kBT) in the most positive regions and deep red (−15kBT) in the most negative, with linear interpolation for values in between. Residues known to be involved in binding are shown.
Binding of ssDNA in the (SSB)65 binding mode would require bridging these two binding surfaces. By crossing over from one side to the other, ssDNA would come into proximity of the base of loop L45 and might involve the loop in binding. Previous structural studies on proteins containing an OB fold used for ssDNA binding have pointed out the importance of the L45 loop. In the structure of tRNA synthase bound to tRNAAsp or in that of the ssDNA binding domain of replication protein A bound to ssDNA, loop L45 bends over the DNA and contributes significant surfaces to binding (53, 54). In E. coli SSB, the L45 loop may play a similar role. It contains Trp-88, and mutations at Trp-88 can affect the relative stabilities of the different SSB–ssDNA binding modes (46). On the other hand, L45 may also play an additional role in E. coli SSB: this loop is an essential component of the L45-mediated dimer–dimer interface observed in the crystal. Therefore, L45 may also be involved in stabilizing cooperative interactions between SSB tetramers bound to long ssDNA. In fact, it may be that loop L45 alternates between interactions with ssDNA or with a second tetramer, depending on the particular single-stranded polynucleotide binding mode. More specifically, in the (SSB)65 binding mode, loop L45 might be involved in ssDNA binding and therefore would not be available to form the L45-mediated dimer–dimer interface. As a consequence, unlimited cooperative interactions between tetramers could not occur. Conversely, in the (SSB)35 binding mode, loop L45 might not be involved in ssDNA binding and therefore could participate in the formation of a L45-mediated dimer–dimer interface: unlimited cooperative interactions between tetramers could then occur.
Visual inspection of the figures in ref. 20 clearly indicate that the structure of SSBC resembles closely that of hmtSSB, with all important amino acids appearing to map to equivalent regions in each structure. Yang et al. (20) propose a model for binding of ssDNA to the hmtSSB tetramer where ssDNA binds across the tetramer. Although we suggest that E. coli SSB may bind ssDNA in a similar way, the actual path for ssDNA binding is still unclear and must await the determination of meaningful ssDNA/E. coli SSB or ssDNA/hmtSSB complex structures. Interestingly, the cocrystal structure of replication protein A (RPA) with ssDNA (53) suggests that binding of ssDNA to homotetrameric SSBs might involve the L12 and L45 loops. When the first SSB module of RPA (residues 183–275) is superimposed onto the SSB monomer, residues Phe-60 (between strand 3 and the helix) and Arg-21 (in strand 1′) are within hydrogen bonding distance of the DNA. However, it is not known whether Arg-21 or any of the residues in the region of the L12 loop of E. coli SSB is involved in binding. Furthermore, in the RPA/SSB superimposition, Trp-54 of E. coli SSB, a residue known to be involved in ssDNA binding, is more than 14 Å away from the DNA. Therefore no definitive conclusion can be drawn from a comparative study of RPA and E. coli SSB.
Acknowledgments
We thank D. J. Leahy (Johns Hopkins University) and T. C. Terwilliger (Los Alamos National Laboratory) for advice on MAD phasing; M. Ludwig (University of Michigan) for advice on NCS averaging; W. van Zante and M. Ferrari for protein purification; S. Korolev for help in data collection; R. D. Porter and L. E. Carlini (Pennsylvania State University) for generously providing information on second site suppressors prior to publication; the staff of Beamline 7.1 at Stanford Synchrotron Radiation Laboratory for assistance during synchrotron collection; C. Ogata and the staff of Beamline X-4A at National Synchrotron Light Source for assistance during synchrotron data collection; an anonymous reviewer for drawing attention to the proper sequence alignment of hmtSSB and Eco SSB, thereby allowing the assignment of Met-23; A. Edwards for the coordinates of RPA bound to ssDNA. Beamline X-4A at the National Synchrotron Light Source is supported by the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grants GM54033 to G.W. and GM30498 to T.M.L.
ABBREVIATIONS
- SSB
single-stranded DNA-binding protein
- MAD
multiwavelength anomalous dispersion
- hmtSSB
human mitochondrial SSB
- ssDNA
single-stranded DNA
- OB
oligomer binding
Footnotes
Data deposition: The atomic coordinates have been deposited at the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB entry code 1KAW).
References
- 1.Sigal N, Delius H, Kornberg T, Gefter M L, Alberts B M. Proc Natl Acad Sci USA. 1972;69:3537–3541. doi: 10.1073/pnas.69.12.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B M, Frey L. Nature (London) 1970;227:1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 3.Meyer R R, Laine P S. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chase J W, Williams K R. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 5.Lohman T M, Ferrari M E. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 6.Lohman T M, Bujalowski W, Overman L B. Trends Biochem Sci. 1988;13:250–255. [PubMed] [Google Scholar]
- 7.Williams K R, Murphy J B, Chase J W. J Biol Chem. 1984;259:11804–11811. [PubMed] [Google Scholar]
- 8.Lohman T M, Overman L B. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 9.Bujalowski W, Lohman T M. Biochemistry. 1986;25:7799–7802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 10.Griffith J D, Harris L D, Register J. Cold Spring Harbor Symp Quant Biol. 1984;49:553–559. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 11.Bujalowski W, Lohman T M. J Mol Biol. 1989;207:249–268. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 12.Bujalowski W, Lohman T M. J Mol Biol. 1989;207:269–288. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 13.Krauss G, Sindermann H, Schomburg U, Maass G. Biochemistry. 1981;20:5346–5352. doi: 10.1021/bi00521a040. [DOI] [PubMed] [Google Scholar]
- 14.Chrysogelos S, Griffith J. Proc Natl Acad Sci USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohman T M, Overman L B, Datta S. J Mol Biol. 1986;187:603–615. doi: 10.1016/0022-2836(86)90338-4. [DOI] [PubMed] [Google Scholar]
- 16.Bujalowski W, Lohman T M. J Mol Biol. 1987;195:897–907. doi: 10.1016/0022-2836(87)90493-1. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Williams K R, Chase J W, Rupp W D. Proc Natl Acad Sci USA. 1981;78:4274–4278. doi: 10.1073/pnas.78.7.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams K R, Spicer E K, LoPresti M B, Guggenheimer R A, Chase J W. J Biol Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 19.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Eur J Biochem. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang C, Curth U, Urbanke C, Kang C. Nat Struct Biol. 1997;4:153–157. doi: 10.1038/nsb0297-153. [DOI] [PubMed] [Google Scholar]
- 21.Ollis D, Brick P, Abdel-Meguid S S, Murthy K, Chase J W, Steitz T A. J Mol Biol. 1983;170:797–801. doi: 10.1016/s0022-2836(83)80134-x. [DOI] [PubMed] [Google Scholar]
- 22.Monzingo A F, Christiansen C. J Mol Biol. 1983;170:801. doi: 10.1016/s0022-2836(83)80135-1. [DOI] [PubMed] [Google Scholar]
- 23.Hilgenfeld R, Saenger W, Schomburg U, Krauss G. FEBS Lett. 1984;170:143–146. [Google Scholar]
- 24.Ng J D, McPherson A. J Biomol Struct Dynam. 1989;6:1071–1076. doi: 10.1080/07391102.1989.10506537. [DOI] [PubMed] [Google Scholar]
- 25.Thorn J M, Carr P D, Chase J W, Dixon N E, Ollis D. J Mol Biol. 1994;240:396–399. doi: 10.1006/jmbi.1994.1453. [DOI] [PubMed] [Google Scholar]
- 26.Lohman T M, Green J M, Beyer R S. Biochemistry. 1986;25:21–25. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Hendrickson W A, Kalman E T, Crouch R J. J Biol Chem. 1990;265:13553–13559. [PubMed] [Google Scholar]
- 28.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyers L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Lab.; 1993. pp. 56–62. [Google Scholar]
- 29.Terwilliger T C, Eisenberg D. Acta Crystallogr A. 1983;39:813–817. [Google Scholar]
- 30.Terwilliger T C. Acta Crystallogr D. 1994;50:11–16. doi: 10.1107/S0907444993008224. [DOI] [PubMed] [Google Scholar]
- 31.Hall T M T, Porter J A, Beachy P A, Leahy D J. Nature (London) 1995;378:212–216. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- 32.Terwilliger T C, Kim S-H, Eisenberg D. Acta Crystallogr A. 1987;43:1–5. [Google Scholar]
- 33.Terwilliger T C. Acta Crystallogr D. 1994;50:17–23. doi: 10.1107/S0907444993008236. [DOI] [PubMed] [Google Scholar]
- 34.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Jones T A, Thirup S. EMBO J. 1986;5:819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read R. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 37.Brünger A. x-plor Manual. New Haven, CT: Yale Univ.; 1988. , Version 2.2. [Google Scholar]
- 38.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 39.Brünger A T. J Mol Biol. 1988;203:803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran G N, Sasisekharan V. Adv Protein Chem. 1968;23:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 41.Murzin A G. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 43.Janin J, Chothia C. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 44.Bujalowski W, Lohman T M. J Biol Chem. 1991;266:1616–1626. [PubMed] [Google Scholar]
- 45.Carlini L E. Ph.D. thesis. University Park: Pennsylvania State Univ.; 1994. [Google Scholar]
- 46.Curth U, Greipel J, Urbanke C, Maass G. Biochemistry. 1993;32:2585–2591. doi: 10.1021/bi00061a016. [DOI] [PubMed] [Google Scholar]
- 47.Anderson R A, Coleman J E. Biochemistry. 1975;14:5485–5491. doi: 10.1021/bi00696a017. [DOI] [PubMed] [Google Scholar]
- 48.Overman L B, Bujalowski W, Lohman T M. Biochemistry. 1988;27:456–471. doi: 10.1021/bi00401a067. [DOI] [PubMed] [Google Scholar]
- 49.Khamis M I, Casas-Finet J R, Maki A H, Murphy J B, Chase J W. J Biol Chem. 1987;262:10938–10945. [PubMed] [Google Scholar]
- 50.Casas-Finet J R, Khamis M I, Maki A H, Chase J W. FEBS Lett. 1987;220:347–352. doi: 10.1016/0014-5793(87)80844-x. [DOI] [PubMed] [Google Scholar]
- 51.Merrill B M, Williams K R, Chase J W, Konigsberg W H. J Biol Chem. 1984;259:10850–10856. [PubMed] [Google Scholar]
- 52.Nicholls A, Sharp K A, Honig B. Protein Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 53.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Nature (London) 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 54.Ruff M, Krishnaswany S, Boeglin M, Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry J C, Moras D. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]