Abstract
The purpose of this study was to understand the characteristics of PDEF protein expression in breast and prostate cancer progression. A polyclonal antibody specific to PDEF was raised and reacted with tissue microarrays consisting of benign breast, in situ ductal, invasive ductal and invasive lobular breast carcinomas. The antibody was also reacted with tissue microarrays including benign prostate, prostate intra-epithelial neoplasias and prostate carcinomas. Increased expression of PDEF was identified in 18%, 50%, 46% and 51% of benign breast tissues, intraductal, invasive ductal and invasive lobular carcinomas, respectively. Importantly, in matched samples of benign breast versus tumor, 90% showed higher expression of PDEF in the tumor tissue. Moreover, in invasive breast carcinomas, increased PDEF expression tended to correlate with Her2/neu over expression. Increased expression of PDEF was also found in 27%, 33% and 40% of benign prostate tissues, PIN samples and prostate adenocarcinomas, respectively. Again, in matching samples of cancer versus benign and cancer versus PIN, 68% and 70% respectively showed increased expression in the malignant tissue. Moreover, PDEF was found to be more highly expressed in tumors with intermediate or high Gleason score compared to low grade tumors (P<0.01). Additionally, R1881 treatment induced PDEF expression in the LNCaP prostate tumor cell line, suggesting regulation of PDEF by androgens in vivo. Together, these results for the first time show frequent increased expression of PDEF protein in breast and prostate tumors and support a role for PDEF in breast and prostate cancer progression.
Keywords: PDEF, Breast cancer, Prostate cancer, Gleason score, R1881, Tumor marker and target
Introduction
Breast cancer is the most common malignancy in women, as about a million new cases of this cancer are diagnosed world-wide each year, and 375,000 women die from it (1). Although early detection through screening mammography has increased the proportion of in situ and early stage breast cancers that have excellent prognosis, mortality rates from recurrent and late stage breast cancers have not declined significantly (2). Consequently, conventional therapies including surgery, radiation and chemotherapy need to be supplemented with novel therapies that translate into significant improvement in the clinical outcome for most breast cancer patients.
Similarly, prostate cancer is the most commonly diagnosed malignancy in men, with about 220,000 new cases of prostate cancer diagnosed each year in the U.S. alone. Of these, approximately two-thirds are treated by surgery or radiation therapy, and 40% of the treated men will eventually relapse, as characterized by rising levels of prostate specific antigen (3, 4). Relapsed advanced and metastatic prostate cancer remains the primary cause of death from this cancer, since current systemic hormonal and chemotherapy approaches are only marginally successful (5). Consequently there is an urgent need for novel therapies to treat advanced prostate cancer.
A targeted approach to cancer therapy has attracted considerable attention following its success in chronic myelogenous leukemia (6). Among the major solid tumors, breast and prostate cancers are perhaps unique in having a history of targeted therapies. Thus, patients with estrogen receptor (ER)-positive and Her2/neu-over expressing breast tumors or androgen receptor (AR)-positive prostate tumors are routinely treated with specific inhibitors that result in consistent remissions in the setting of early disease (7-9). Similarly, a less common neoplasm, gastrointestinal stromal tumor, also shows high susceptibility to targeted inhibition of the activity of mutated KIT oncogene (10). With such promise of targeted cancer therapies, much effort in the past decade has focused on identifying useful novel cancer targets. The need for novel targets is particularly acute for breast and prostate cancers because of the immense cancer burden world-wide and because the present repertoire of useful breast and prostate cancer targets is limited. To that end, molecules that play a role in breast and prostate cancer progression are likely to be useful targets against these cancers.
Prostate derived Ets factor (PDEF) belongs to the Ets family of transcription factors that play an important role in normal as well as neoplastic development (11-15). However, despite such promise, our knowledge about the characteristics of PDEF expression in human cancer remains limited. To address this deficiency, we generated a polyclonal antibody to an N-terminal segment of PDEF that is unique to PDEF. By using this antibody, tissue microarrays (TMAs) consisting of a large number of benign breast and prostate tissues and from various stages of breast and prostate neoplasia were screened for PDEF expression. Our results show frequent increased expression of PDEF protein in both breast and prostate carcinomas compared to benign tissues. Further, PDEF expression tends to associate with Her2/neu over expression in invasive breast carcinomas and shows significant correlation with intermediate to high grade prostate carcinomas. Together, these results provide novel insights into the characteristics of PDEF protein expression in breast and prostate cancers and support its role in the progression of these cancers.
2. Materials and methods
2.1. Cell lines and antibodies
MCF-7, SKBR3, U937 and Hela tumor cell lines were obtained from colleagues at Roswell Park Cancer Institute. These cell lines were grown in DMEM medium supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 0.1 μg/ml streptomycin in a 5% CO2 incubator. The actin monoclonal antibody and HRP- conjugated goat anti-rabbit antibody were purchased from Sigma (St. Louis, MO).
2.2. Generation of polyclonal antibodies against an N-terminal segment of PDEF
The full-length open reading frame of PDEF cDNA (PDEF-full-length) and the cDNA encoding N-terminal 1 to 104 amino acids (PDEF-1-104) were PCR-amplified and subcloned into the bacterial expression vector pET15b (Novagen, Madison, WI) at the Nde I site. After confirmation of the orientation and sequence, E. coli BL21(DE3) cells were transformed by the resultant pET15b-PDEF-full-length or pET15b-PDEF-1-104 plasmids. The cells were induced by 1mM IPTG and PDEF protein was purified from bacterial lysates by affinity chromatography on a Ni-NTA column. Further purification on MonoQ column (Amersham, Piscataway, NJ) provided pure preparations of the PDEF-full-length and PDEF-1-104 peptides. The purified PDEF-1-104 peptide was used for production of polyclonal antibodies in rabbits. Briefly, two New Zealand white rabbits each were immunized by intradermal injection of 150 μg of PDEF-1-104 protein emulsified in complete Freund's adjuvant. This was followed by three booster injections with 150 μg protein in incomplete Freund's adjuvant at two-week interval and the 4th injection at 6 weeks following the third boost. Serum was collected and tested for reactivity by ELISA and Western blot assays using purified PDEF protein as an antigen. PDEF antibodies were further purified on a PDEF-full-length protein-immobilized affinity column (Pierce, Rockford, IL).
2.3. Western blot analysis
Cells from breast and non-breast tumor cell lines were lysed in RIPA buffer containing 50mM Tris-HCl, pH 7.5, 150 mM NaCl, 1mM PMSF, 1mM EDTA, 5 μg/ml Aprotinin, 5 μg/ml Leupeptin, 1% Triton x-100, 1% Sodium deoxycholate and 0.1% SDS. The protein concentration of the cell lysates was determined using a BCA kit (Pierce). 50 μg protein from each cell line was loaded onto 12% SDS-PAGE and following electrophoresis protein was transferred to nitrocellulose membrane. Western blot analysis of PDEF and actin expression was performed using the polyclonal rabbit anti-PDEF-1-104 antibody described above and the actin antibody respectively. Signals were detected using a HRPL kit (National Diagnostics/LPS, Rochester, NY) or a Chemo-luminescent Reagent Plus kit (Perkin Elmer) and visualized by autoradiography.
2.4. Tissue sections and tissue microarrays
Institutional Review Board approval was obtained prior to initiation of this study. Briefly, five µm paraffin sections from normal human tissues were prepared from excess archival tissues in the surgical pathology files at our institute. Tissue micro arrays (TMAs) were constructed from a cohort of 350 formalin fixed, paraffin embedded breast carcinomas treated at Roswell Park Cancer Institute between 1993 and 2001. This cohort was enriched for carcinomas with lobular differentiation (n=117). Only cases with at least two tumor blocks were included. Two 1 mm cores were transferred from each tumor to recipient blocks, using a manual arrayer from Beecher Instruments. For each case, we sampled invasive carcinoma and morphologically benign breast tissue. If available, DCIS was also sampled and transferred to different recipient blocks.
TMAs were also constructed, in duplicate, from a cohort of prostate cancer patients (n=329) treated at Roswell Park Cancer Institute. The corresponding paraffin blocks were retrieved from the surgical pathology files. For each case, we sampled the primary invasive prostate carcinoma and morphologically benign tissue from the same specimen. Also, PIN lesions from the same cohort were sampled and included in separate TMAs. The Gleason scores (GS) of the invasive prostate carcinomas were derived from pathology reports and categorized as low (GS2-5), intermediate (GS6-7) and high (GS8-10) (16).
Due to technical limitations, not all tissue cores were evaluable for PDEF expression by immunohistochemistry. The breast TMAs, in particular, were characterized by variable numbers of missing or uninterpretable cores. The numbers of evaluable samples in each category are listed in the Results section (see later).
2.5. Immunohistochemical assay for PDEF
The paraffin sections of the normal tissues and of the TMAs were dewaxed and heated to 95-100°C for 20 min in high pH antigen retrieval buffer (Dako, Carpinteria, CA). The slides were then reacted with affinity purified rabbit anti-PDEF antibody at a 1:200 dilution at 4°C overnight. The detection reaction utilized the rabbit Envision kit from Dako. Diaminobenzidine (DAB) was used as chromogen, and hematoxylin as counterstain. Paraffin-embedded MCF-7 cell buttons were used as positive controls.
2.6. Scoring of PDEF expression
Both the average nuclear staining intensity and the percentage of stained cells were evaluated. Epithelial tissues containing a minimum of 10% cells with strong (3+) staining or 15% cells with moderate (2+) staining or 30% cells with weak (1+) staining were considered positive for PDEF expression. All other tissues including those that showed staining below these thresholds or lacked any positively stained cells were scored negative for PDEF expression.
2.7. Induction of PDEF expression in LNCaP cells in vitro by DHT analogue R1881
1×106 cells/well from LNCaP prostate tumor cell line were incubated in a 6-well plate in RPMI medium containing charcoal stripped fetal bovine serum for 5 days. Following this, cells were treated with various concentrations of R1881 ranging from 0 to 100 nM by using appropriate amounts from a freshly prepared stock solution of R1881 in ethanol. The cells were incubated in the presence of R1881 for 24 hr, following which total RNA and protein were isolated. PDEF expression in these samples was then analyzed by RT/PCR and Western blotting. For RT/PCR 1 µg of total RNA was used and for Western blotting 40 µg of total protein was used.
3. Results
3.1. Anti-PDEF antibody shows specific reactivity with PDEF
The Blast homology search for PDEF protein sequence showed significant homology (45 to 55% amino acid residue identity) with the C-terminal DNA binding domain of other Ets factors (11). Further, D-Ets-4 (the putative Drosophila homologue of PDEF) showed 40% identity with PDEF that spans amino acid residues 104 through 335 (data not shown). In contrast, the N-terminal segment of PDEF comprising residues 1 to 104 revealed homology only to PDEF, and therefore was selected for producing polyclonal antibodies in rabbits. Following affinity purification, the antibody was tested for specificity by Western blot assay and by immunohistochemical staining of cells from MCF-7 breast tumor cell line and from tissue sections from primary breast and prostate tumors. The data are shown in Figure 1.
Figure 1.
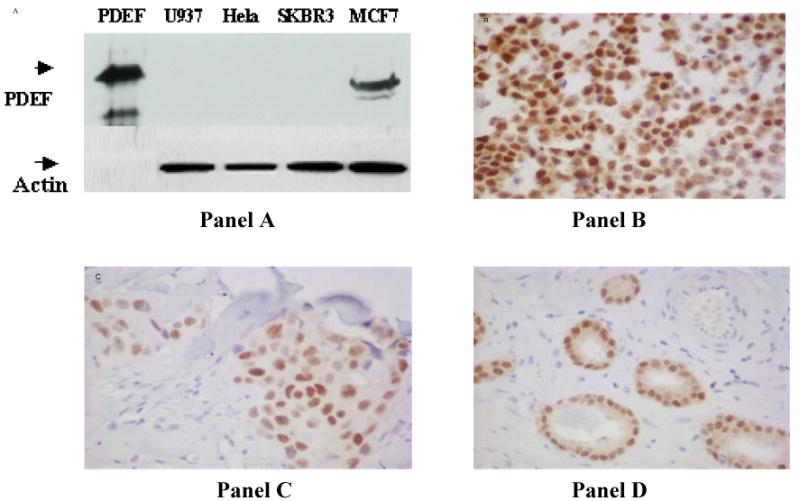
Testing the specificity of anti-PDEF antibodies by Western blotting and immunohistochemistry. Panel A shows analysis of PDEF protein expression in breast and non-breast tumor cell lines by Western blotting. Equal amounts of total protein from breast tumor cell lines MCF-7, SKBR3 and from non-breast tumor cell lines Hela and U937 were run on 12% SDS PAGE and transferred to nitrocellulose membrane. The upper-half of this panel shows the result of probing the blot with anti-PDEF antibodies and the lower-half shows a similar blot following probing with anti-actin antibody. The Panels B, C and D respectively show specific nuclear staining of PDEF protein in the epithelial tumor cells from MCF-7 breast tumor cell line and in tissue section from a primary breast carcinoma and a primary prostate carcinoma.
As shown in Panel A of Figure 1, anti-PDEF antibody reacted specifically with a 46 k.d. band in the MCF-7 breast tumor cell line, but not with the SKBR3 breast tumor cell line or the non-breast tumor cell lines Hela and U937. Also, the antibody reacted strongly with recombinant PDEF protein run as control. Further, this antibody showed specific nuclear staining of tumor cells in the MCF-7 breast tumor cell line (Figure 1, panel B), that was eliminated by prior incubation of the antibody with recombinant PDEF-1-104 peptide (data not shown). Moreover, screening of the tissue sections from a primary breast carcinoma (Figure 1, Panel C) and a primary prostate carcinoma (Figure 1, Panel D) showed specific staining of the epithelial tumor cells, a result consistent with epithelial cell specific expression of PDEF mRNA (11) and of Pse (prostate specific Ets, the mouse homologue of PDEF) mRNA (17). These results together show the specificity of the anti-PDEF antibody as well as its suitability for use in immunohistochemical analysis of PDEF expression in primary tumors.
3.2. Screening of normal human tissues further validates PDEF specificity of our antibody
Previous work on PDEF mRNA expression in normal human tissues reported its restricted expression to normal prostate and trachea (11, 18). To determine whether PDEF protein expression corresponds with PDEF mRNA expression, we screened tissue sections from a panel of normal human tissues for PDEF protein expression using our antibody. Our data show that strong expression of PDEF protein was present in the normal prostate tissue (Figure 2, panel A) and a relatively weaker expression in the normal bronchus tissue (Figure 2, panel B). Other normal human tissues including brain, heart, kidney, liver, lung, lymph nodes, ovary, pancreas, placenta, skeletal muscle, skin, spleen, stomach and thymus stained negative for PDEF protein expression (data not shown). These results are in agreement with the corresponding data on PDEF mRNA expression in the respective tissues (11, 18) and further establish the specificity of our anti-PDEF antibody used in this study.
Figure 2.
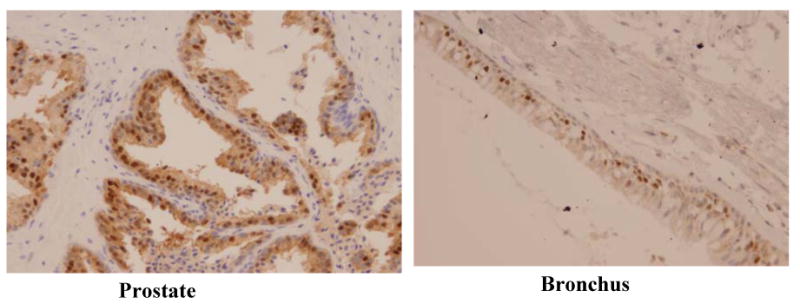
PDEF protein expression in normal human tissues. Photomicrographs show strong expression of PDEF in normal prostate tissue and relatively weaker expression in the normal bronchus tissue.
3.3. PDEF expression is increased in progression from benign breast to DCIS and increased expression is maintained in invasive carcinomas
Using the above antibody, and the scoring criteria described in Materials and Methods, TMAs of breast tissues were screened for PDEF expression. The data from this screening are presented in Figure 3 and are summarized below.
Figure 3.
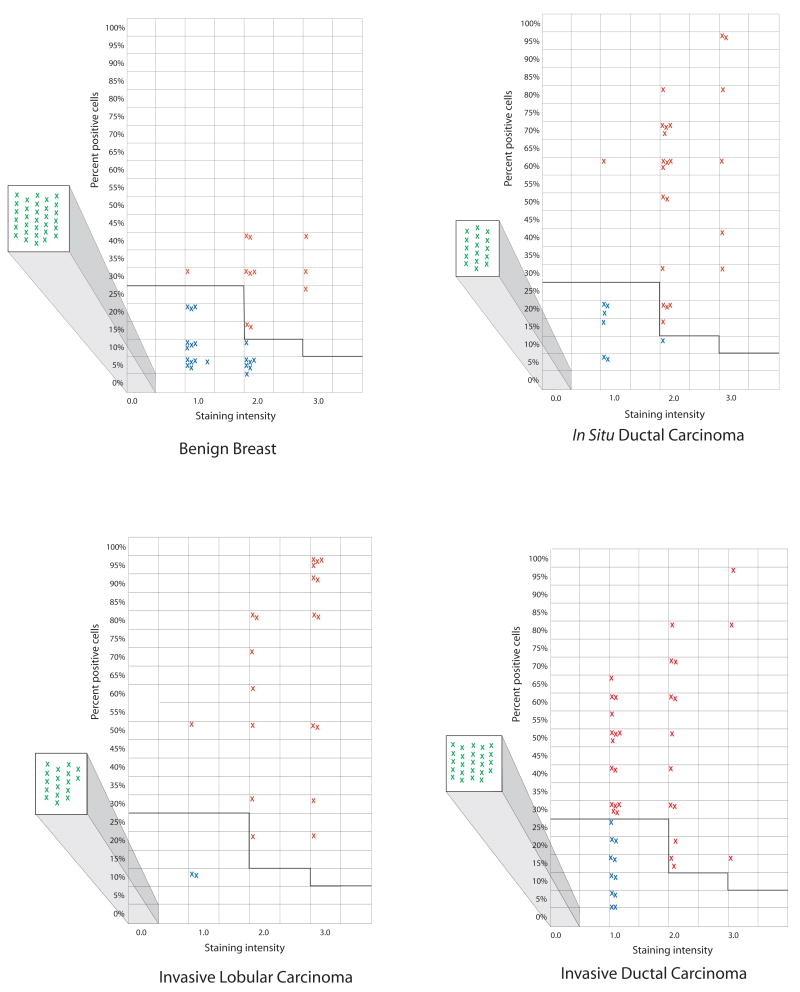
PDEF expression in benign breast tissues, intraductal carcinomas (DCIS), invasive ductal carcinomas and invasive lobular carcinomas. Negative samples (all tumor cells lacking staining) are shown as green crosses inside the squares projecting out to the left of each graph. The samples showing PDEF expression below thresholds are shown as blue crosses in the main graphs. The red crosses in the graphs represent individual samples that stained above the selected thresholds and therefore scored positive for PDEF expression. The x-axis in these graphs shows different levels of staining intensity i.e. 1+, 2+ and 3+. The y-axis shows percent of positively staining cells in a given tumor sample.
In benign breast tissue samples, 18% (11 of 62) scored positive for PDEF expression. Of the remaining samples 20 of 62 (32%) showed expression levels below the selected thresholds and 31 of 62 (50%) lacked any detectable PDEF expression.
For DCIS, 50% (23 of 46) of the samples scored positive for PDEF expression, another 7 (15%) stained below the thresholds and the remaining 16 of 46 (35%) showed no detectable staining.
A similar analysis of invasive ductal carcinoma (IDC) showed that 46% (30 of 65) of the invasive tumors were positive for PDEF expression. Of the remaining samples 17% (11 of 65) showed staining below the thresholds and 24 of 65 (37%) lacked detectable staining.
In invasive lobular carcinoma (ILC), 51% (20 of 39) of the samples scored positive for PDEF expression, another 5% (2 of 39) stained below the thresholds and the remaining 44% (17 of 39) were negative.
In summary, the above results show that whereas 18% of benign breast tissues scored positive for PDEF, samples from DCIS, IDC and ILC showed a much higher percentage of PDEF positive tumors i.e., 50%, 46% and 51% of the screened samples respectively.
3.4. Increased expression of PDEF in breast tumors in comparison to matched benign breast tissues results from increased percentage and/or increased staining of tumor epithelial cells
The data in Figure 3 show an interesting feature of PDEF expression in benign breast tissues versus breast tumors. Specifically, the PDEF positive benign breast tissues generally contained a lower percentage of PDEF expressing epithelial cells in comparison to tumor samples. Thus, in all of the benign breast tissue samples, < 50% of the epithelial cells stained positive for PDEF. In contrast, 35% of DCIS, 25% of IDC and 41% of ILC contained 50% or more epithelial cells staining positive for PDEF. To test this observation further and to determine the characteristics of PDEF protein expression in matched benign and tumor samples from same patients, PDEF expression was analyzed in 9 such matched pairs of adjacent benign breast and tumor tissues. It was found that in 90% (8 of 9) cases, the tumors showed an increase in the number of PDEF expressing epithelial cells and/or the intensity of staining of such cells. The data are summarized in Table 1A and representative photomicrographs are shown in Figure 4.
Table 1.
| Table 1A. PDEF expression in matched samples of benign breast and tumor tissues. | |||
| Benign/ Tumor pair number. | PDEF expression characteristics (Percent positive cells, intensity of staining). | ||
| Benign | Tumor | ||
| 1 | 0 | 40%, 1+ | |
| 2 | 0 | 20%, 2+ | |
| 3 | 40%, 2+ | 60%, 2+ | |
| 4 | 30%, 2+ | 5%, 1+ | |
| 5 | 5%, 1+ | 30%, 3+ | |
| 6 | 5%, 1+ | 20%, 2+ | |
| 7 | 0 | 80%, 2+ | |
| 8 | 15%, 1+ | 65%, 1+ | |
| 9 | 0 | 60%, 2+ | |
| Table 1B. PDEF expression in invasive ductal carcinoma in relation to other prognostic markers. | |||
| Marker Status | PDEF-positive | PDEF-negative | P-value |
| Her-2/neu-positive | 15 | 14 | 0.1478 |
| Her-2/neu-negative | 8 | 17 | |
| ER-positive | 24 | 26 | 0.2046 |
| ER-negative | 4 | 10 | |
| PR-positive | 18 | 21 | 0.7097 |
| PR-negative | 10 | 14 | |
| Tumor grades I and II | 17 | 20 | 0.6652 |
| Tumor grade III | 11 | 16 | |
| Lymph node-positive | 8 | 9 | 0.8371 |
| Lymph node-negative | 9 | 10 | |
Figure 4.
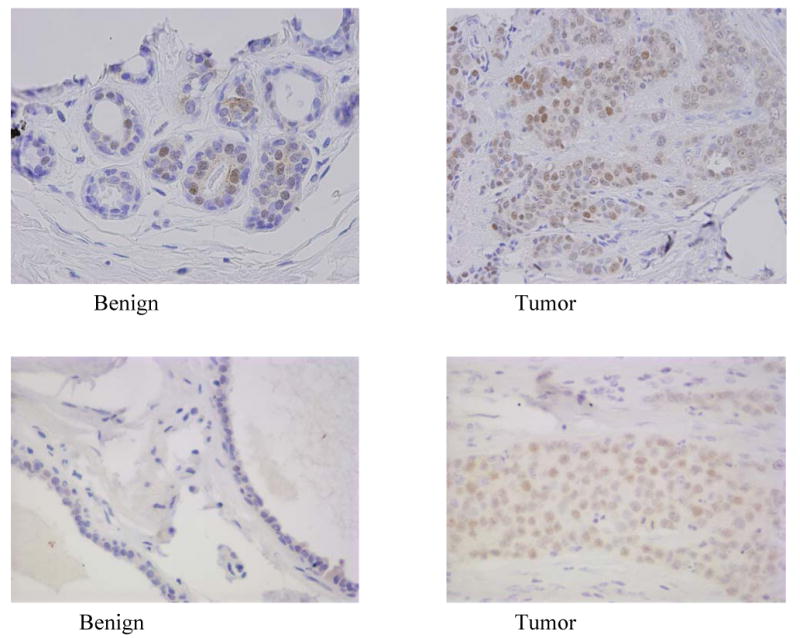
PDEF expression in matched pairs of benign breast and breast tumor tissues. The upper and lower pairs of photomicrographs show representative PDEF staining in matched samples of adjacent benign breast versus breast tumor tissues from two cases. Increased staining in tumor tissues is seen in comparison to benign tissues.
3.5. PDEF expression tends to correlate with Her2/neu over expression and shows lack of correlation with ER and PR expression, tumor grade or lymph node metastases
To determine any correlation of PDEF expression with other known prognostic markers in breast cancer, PDEF-positive and PDEF-negative invasive breast carcinomas were analyzed for Her2/neu, ER and PR expression, tumor grade and for lymph node metastases status obtained from the pathology reports of those patients. The data are shown in Table 1B. Briefly, the Her2/neu-positive tumors were twice as likely to be positive for PDEF expression as Her2/neu-negative tumors. Aside from this tendency, positive PDEF expression did not correlate with ER, PR expression status, tumor grade or presence or absence of lymph node metastases.
3.6. Progressively increasing fractions of benign prostate tissue, PIN and prostate carcinoma samples score positive for PDEF expression
Using the above antibody and the scoring criteria described in Materials and Methods, TMAs of prostate tissues were also stained and scored for PDEF expression. The data are presented in Figure 5 and are described below.
Figure 5.
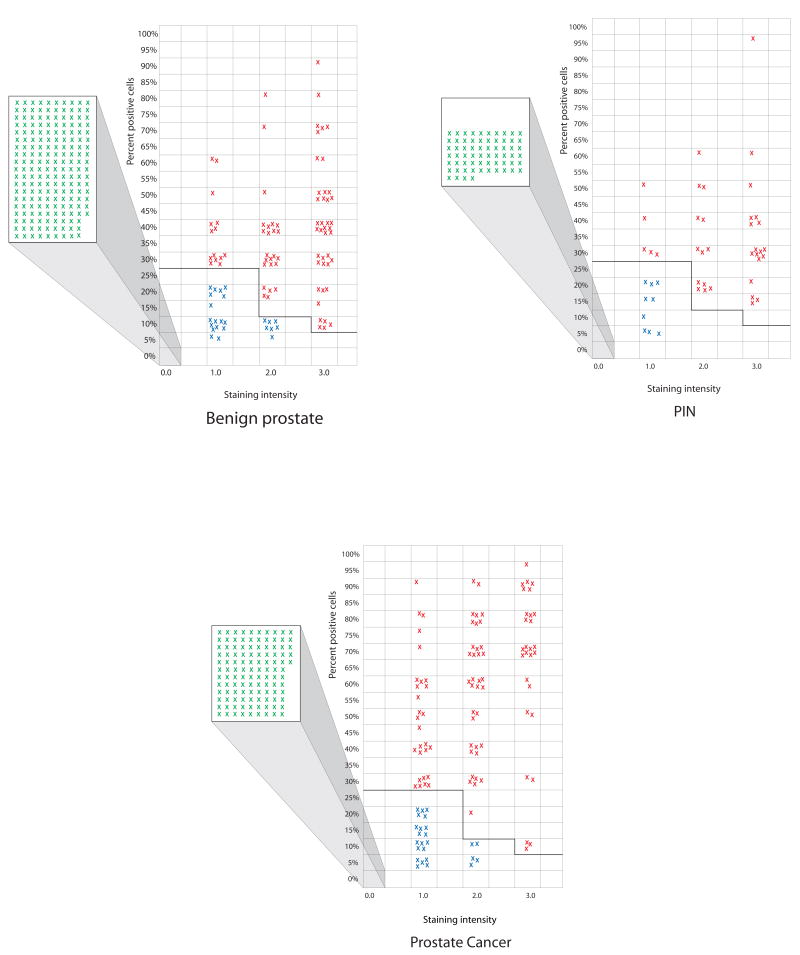
PDEF expression data in samples from benign prostate tissues, prostate intraepithelial neoplasias (PIN) and prostate carcinomas. Negative samples (all tumor cells lacking staining) are shown as green crosses inside the squares projecting out to the left of each graph. The samples showing PDEF expression below thresholds are shown as blue crosses in the main graphs. The red crosses in the graphs represent individual samples that stained above the selected thresholds and therefore scored positive for PDEF expression. The x-axis in these graphs shows different levels of staining intensity i.e. 1+, 2+ and 3+. The y-axis shows percent of positively staining cells in a given tumor sample.
In the benign prostate tissues from cancer patients 27% (79 of 290) scored positive for PDEF expression. Of the remaining samples, 24 of 290 (9%) stained below the selected thresholds and another 187 of 290 (64%) lacked detectable PDEF expression.
For PIN samples, 36 of 109 (33%) scored positive for PDEF expression. Of the remaining, 9 of 109 (8%) stained below the thresholds and 64 out of 109 (59%) showed no detectable staining.
A similar analysis of prostate carcinomas revealed that 40% (92 of 230) of the invasive carcinomas were positive for PDEF expression, another 11% (25 of 230) showed staining below thresholds and remaining 113 out of 230 (49%) samples lacked detectable staining.
In summary, the above results support the following points: i) increasingly higher percentage of samples scored positive for PDEF expression in progression from benign prostate tissues (27% of samples positive), to PIN lesions (33% of samples positive) and to prostate carcinomas (40% of samples positive); ii) prostate carcinomas generally showed a higher percentage of cells staining positive for PDEF in comparison to PIN or benign tissue samples.
3.7. Analysis of PDEF expression in matched samples of cancer versus benign, cancer versus PIN and PIN versus benign showed frequent increased PDEF expression in prostate cancer
To further understand the characteristics of PDEF protein expression in benign prostate and prostate carcinomas, PDEF expression was analyzed in matched samples of prostate cancer and adjacent benign tissue or PIN. Overall, this analysis included 101 matched samples of cancer versus (v/s) benign, 41 matched samples of cancer v/s PIN and 45 matched samples of PIN v/s benign. The data are compiled in Table 2. As shown in this Table, in 68% (68 of 101) of the matched cancer v/s benign pairs, cancer specimens expressed higher levels of PDEF than the adjacent benign prostate tissues. Similarly, in 70% (28 of 41) of the matched cancer v/s PIN pairs, cancer showed higher PDEF expression than PIN. In contrast, in PIN versus benign comparison, only 42% (19 of 45) PIN lesions showed higher PDEF expression than the matched benign glandular tissues.
Table 2. PDEF expression in prostate tissue: matched samples of cancer v/s benign, Cancer v/s PIN and PIN v/s benign.
| Type of Match X v/s Y | Number of matched samples | Number of samples with more* PDEF in X | Number of samples with more* PDEF in Y | P value |
|---|---|---|---|---|
| Cancer v/s benign | 101 | 68 (68%) | 33 (32%) | 0.0006 |
| Cancer v/s PIN | 41 | 28 (70%) | 13 (30%) | 0.0275 |
| PIN v/s benign | 45 | 19 (42%) | 26 (58%) | 0.3713 |
More PDEF reflects higher percent of PDEF-positive epithelial cells or higher intensity of staining or both.
In summary, the combined data from Figure 5 and Table 2 show that PDEF expression is frequently increased in the progression from benign prostate to cancer and from PIN to cancer. In contrast, there appeared to be no significant change in PDEF expression in progression from benign prostate to PIN.
3.8 Correlation of PDEF expression with Gleason score
To determine any correlation of PDEF expression with Gleason Score (GS), we assigned low, intermediate or high GS to prostate carcinomas according to the criteria outlined in Materials and Methods, and analyzed any relationship between their GS and PDEF expression status. We found that prostate cancers with intermediate GS showed the highest correlation with PDEF expression i.e., 77/159 (48%) tumors positive for PDEF. In contrast, 24% (11/45) of high GS but only 15% (4/26) of low GS prostate carcinomas were positive for PDEF. Combining intermediate and high GS tumors as one group (n=204) and comparing them with low GS tumors as another group (n=26), we found that 43% (88/204) of intermediate to high grade v/s 15% (4/26) of low grade prostate carcinomas were positive for PDEF, which was statistically significant (P<0.01).
3.9 PDEF expression is induced by DHT analogue R1881 in the LNCaP prostate tumor cell line
The data in Figure 5 show that the vast majority (73%) of benign prostate tissue samples from prostate cancer patients showed little PDEF expression or expression below the selected thresholds. These results were contrary to our expectation since PDEF is a prostate associated transcription factor, and therefore, should be expressed in benign prostate tissues. To understand this apparent discrepancy we hypothesized that PDEF expression may be regulated by androgens in vivo such that decline in androgen levels in aging prostate cancer patients would result in the decrease or loss of PDEF expression in their prostate tissue. Prostate tumors, on the other hand, may use other mechanisms to up regulate their PDEF expression. To test the potential regulation of PDEF by androgens, the LNCaP prostate tumor cell line was treated with various concentrations of DHT analog R1881 in vitro, and PDEF mRNA and protein expression analyzed in R1881 treated and control cells. The data presented in Figure 6 show that both PDEF mRNA and protein expression are induced when LNCaP cells were treated with R1881.
Figure 6.
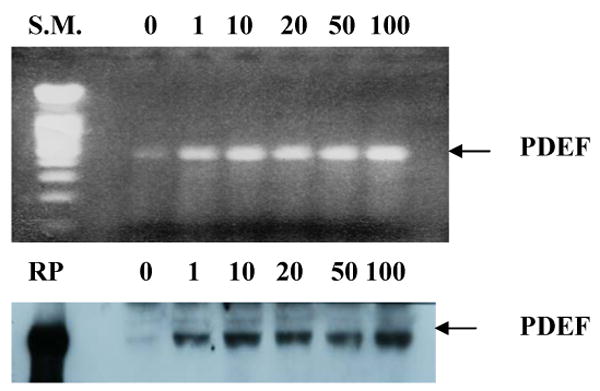
Induction of PDEF expression by DHT analog R1881. LNCaP prostate tumor cells were cultured with various concentrations of R1881 ranging from 1 nM to 100 nM. The upper panel shows PDEF mRNA expression by RT/PCR from control and treated samples. The lower panel shows PDEF protein expression on the same samples by Western blotting using anti-PDEF antibody. SM, in the upper panel, denotes DNA size markers; and RP, in the lower panel, denotes recombinant PDEF protein.
4. Discussion
We previously showed frequent increased expression of PDEF mRNA in breast tumors and its highly restricted expression in normal human tissues (18). Subsequent studies, however, reported loss of PDEF protein expression in the progression from benign breast and prostate tissues to carcinomas (19-21). The present study, for the first time, shows frequent increased expression of PDEF protein in both breast and prostate tumors and these results differ from the conclusions of previous reports (19-21). A potential explanation for differing results in previous studies versus this study may be the small sample sizes analyzed in those studies. Alternatively, since the anti-PDEF antibodies used in two of the three previous studies (19, 20) were generated against the full-length PDEF protein, they carried the potential for cross-reaction with other Ets factors in the immunohistochemical assay. The latter potential stems from: i) the high degree of sequence homology approaching 45 to 55% amino acid sequence identity in the DNA binding domain between PDEF and other Ets factors (11); and ii) the large number of Ets factors that are expressed in normal mammary gland, and several at much higher levels than Pse or PDEF (17, 22). The third study (21) used antibody raised against the same N-terminal segment of PDEF as used in this study, however, the specificity of the immunohistochemical assay for PDEF was not demonstrated in that study. The specificity of our antibody for PDEF was demonstrated rigorously in the experiments shown in Figures 1 and 2 and our results are based on the analysis of a large number of samples.
Our data in Table 1B show that increased PDEF expression tends to associate with Her2/neu over expression in invasive breast carcinomas. A recent report described synergistic co-operation between PDEF and Her2/neu or CSF-1R/CSF-1 in promoting anchorage independent growth, motility and invasiveness in normal and neoplastic breast and non-breast epithelial cell lines (15), suggesting that PDEF collaborates with initiating oncogenic signaling from these receptor tyrosine kinases to enhance tumor progression. In light of the latter understanding, our preliminary finding of an association of increased PDEF expression with Her2/neu over expression is very interesting and needs validation in a larger study.
The increased expression of PDEF in about 50% of both ductal and lobular breast carcinomas (Figure 3) is further interesting since these two histologic types constitute the vast majority of all breast cancers. In contrast, Her2/neu is over expressed in 25 to 30% of invasive ductal carcinomas.
Another noteworthy observation of this study is the lack of or below threshold expression of PDEF in vast majority (73%) of benign prostate tissues from prostate cancer patients (Figure 5). Our hypothesis that declining testosterone levels in aging prostate cancer patients (23-25) may underlie this phenomenon is supported by R1881 regulation of PDEF. Further, our finding that in about 70% of the matched cases, prostate carcinoma show higher PDEF expression than benign or PIN tissues (Table 2) suggests that tumors use novel mechanism(s) to up regulate PDEF expression. In this regard, it is noteworthy that the chromosomal region spanning the 6p21.31 genetic locus, where PDEF gene resides, is frequently amplified in breast and other tumor types (26). Although similar data for prostate tumors is not known, gene amplification remains an interesting potential mechanism to explain tumor-associated increased expression of PDEF.
Analysis of the correlation of PDEF expression with Gleason score in prostate carcinomas showed higher expression levels in intermediate and high grade v/s low grade tumors, supporting the notion that increased expression of PDEF may be associated with a more aggressive phenotype.
Targeting of transcription factors is a well-established paradigm in cancer therapy (27-29). A transcription factor can significantly impact the biology of a tumor by inducing large scale changes in gene expression, some of which may encode cell surface or secreted molecules capable of influencing the behavior of the neighboring tumor and/or stromal cells. This notion is supported by the characteristics of estrogen receptor expression in breast cancer and its implication for endocrine therapy. Thus, even when a small fraction (10%) of the cells of a breast tumor express the estrogen receptor, that tumor is scored positive for ER expression and endocrine therapy shows clinical benefit for the patient (30, 31). In a similar vein, PDEF as an over-expressed transcription factor in breast and prostate cancers may significantly influence the biology of these tumors, and serve as an important marker and target in these cancers.
In conclusion, our work provides the first in-depth study of the characteristics of PDEF protein expression in breast and prostate tumors. The results show frequent increased expression of PDEF in both cancers, a tendency to associate with Her2/neu over expression in breast cancer, significant association with intermediate to high Gleason score in prostate cancer, and potential for androgen independent expression in prostate tumors. Together, these results support a role for PDEF in breast and prostate cancer progression, and as a candidate novel marker and target in these cancers.
Acknowledgments
The authors thank Joseph Brachmann for help with antibody production and Hillary Banas for help with preparation of Figures 3 and 5 of this manuscript. Thanks are also due to Dr. Liaomin Peng for critical review of this manuscript and to Dr. Soldano Ferrone for his support and interest in this work.
This work was supported by a grant CA 86164 from National Cancer Institute and by a grant BC045095 from U.S. Army. The institutional core facilities used in this research were supported by Roswell Park Cancer Center Support Grant P30CA16056. The authors do not have a financial conflict of interest in regard to this study.
Nonstandard Abbreviations
- DCIS
Ductal carcinoma in situ
- DHT
Dihydrotestosterone
- IDC
Invasive ductal carcinoma
- ILC
Invasive lobular carcinoma
- PDEF
Prostate derived Ets factor
- PIN
Prostate intraepithelial neoplasia
- Pse
Prostate specific Ets
- TMA
Tissue microarray
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6:229–39. doi: 10.1186/bcr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chew HK. Medical management of breast cancer: today and tomorrow. Cancer Biother Radiopharm. 2002;17:137–49. doi: 10.1089/108497802753773766. [DOI] [PubMed] [Google Scholar]
- 3.Ward JF, Moul JW. Biochemical recurrence after definitive prostate cancer therapy. Part I: defining and localizing biochemical recurrence of prostate cancer. Curr Opin Urol. 2005;15:181–86. doi: 10.1097/01.mou.0000165552.79416.11. [DOI] [PubMed] [Google Scholar]
- 4.Ward JF, Moul JW. Biochemical recurrence after definitive prostate cancer therapy. Part II: treatment strategies for biochemical recurrence of prostate cancer. Curr Opin Urol. 2005;15:187–95. doi: 10.1097/01.mou.0000165553.17534.e3. [DOI] [PubMed] [Google Scholar]
- 5.Ryan CJ, Small EJ. Progress in detection and treatment of prostate cancer. Curr Opin Oncol. 2005;17:257–60. doi: 10.1097/01.cco.0000155008.37524.8e. [DOI] [PubMed] [Google Scholar]
- 6.O'Dwyer ME, Druker BJ. The role of the tyrosine kinase inhibitor STI571 in the treatment of cancer. Curr Cancer Drug Targets. 2001;1:49–57. doi: 10.2174/1568009013334250. [DOI] [PubMed] [Google Scholar]
- 7.Jordan VC, Brodie AM. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids. 2007;72:7–25. doi: 10.1016/j.steroids.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG, Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas EP, Lingle WL, Klein PM, Ingle JN, Wolmark N. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 9.Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr Relat Cancer. 2004;11:459–76. doi: 10.1677/erc.1.00525. [DOI] [PubMed] [Google Scholar]
- 10.Gold JS, Dematteo RP. Combined surgical and molecular therapy: the gastrointestinal stromal tumor model. Ann Surg. 2006;244:176–84. doi: 10.1097/01.sla.0000218080.94145.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem. 2000;275:1216–25. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 12.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 13.Mimeault M. Structure-function studies of ETS transcription factors. Crit Rev Oncog. 2000;11:227–53. [PubMed] [Google Scholar]
- 14.Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–70. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- 15.Gunawardane RN, Sgroi DC, Wrobel CN, Koh E, Daley GQ, Brugge JS. Novel role for PDEF in epithelial cell migration and invasion. Cancer Res. 2005;65:11572–80. doi: 10.1158/0008-5472.CAN-05-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostofi FK, Sesterhenn I, Davis CJ, Sobin LH. Histological typing of prostate tumours. 2. Berlin; New York: Springer; 2002. p. 115. [Google Scholar]
- 17.Galang CK, Muller WJ, Foos G, Oshima RG, Hauser CA. Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. J Biol Chem. 2004;279:11281–92. doi: 10.1074/jbc.M311887200. [DOI] [PubMed] [Google Scholar]
- 18.Ghadersohi A, Sood AK. Prostate epithelium-derived Ets transcription factor mRNA is overexpressed in human breast tumors and is a candidate breast tumor marker and a breast tumor antigen. Clin Cancer Res. 2001;7:2731–38. [PubMed] [Google Scholar]
- 19.Nozawa M, Yomogida K, Kanno N, Nonomura N, Miki T, Okuyama A, Nishimune Y, Nozaki M. Prostate-specific transcription factor hPSE is translated only in normal prostate epithelial cells. Cancer Res. 2000;60:1348–52. [PubMed] [Google Scholar]
- 20.Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003;63:4626–31. [PubMed] [Google Scholar]
- 21.Ghadersohi A, Pan D, Fayazi Z, Hicks DG, Winston JS, Li F. Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibits breast cancer cell growth in vitro and xenograft tumor formation in vivo. Breast Cancer Res Treat. 2007;102:19–30. doi: 10.1007/s10549-006-9314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Pan Y, Hu J, Albarracin C, Wu Y, Le Dai J. Profile of Ets Gene Expression in Human Breast Carcinoma. Cancer Biol Ther. 2007;6:76–82. doi: 10.4161/cbt.6.1.3551. [DOI] [PubMed] [Google Scholar]
- 23.Veldhuis JD, Keenan DM, Iranmanesh A. Mechanisms of ensemble failure of the male gonadal axis in aging. J Endocrinol Invest. 2005;28:8–13. [PubMed] [Google Scholar]
- 24.Haren MT, Morley JE, Chapman IM, O'Loughlin PD, Wittert GA. Defining ‘relative’ androgen deficiency in aging men: how should testosterone be measured and what are the relationships between androgen levels and physical, sexual and emotional health? Climacteric. 2002;5:15–25. [PubMed] [Google Scholar]
- 25.Hijazi RA, Cunningham GR. Andropause: is androgen replacement therapy indicated for the aging male? Annu Rev Med. 2005;56:117–37. doi: 10.1146/annurev.med.56.082103.104518. [DOI] [PubMed] [Google Scholar]
- 26.Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification and cancer progression. J Clin Pathol. 2007;60:1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libermann TA, Zerbini LF. Targeting transcription factors for cancer gene therapy. Curr Gene Ther. 2006;6:17–33. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Li M, Rayburn ER, Hill DL, Zhang R, Wang H. Oncogenes as novel targets for cancer therapy (part III): transcription factors. Am J Pharmacogenomics. 2005;5:327–38. doi: 10.2165/00129785-200505050-00005. [DOI] [PubMed] [Google Scholar]
- 29.Redell MS, Tweardy DJ. Targeting transcription factors for cancer therapy. Curr Pharm Des. 2005;11:2873–87. doi: 10.2174/1381612054546699. [DOI] [PubMed] [Google Scholar]
- 30.Rhodes A, Jasani B, Balaton AJ, Barnes DM, Miller KD. Frequency of oestrogen and progesterone receptor positivity by immunohistochemical analysis in 7016 breast carcinomas: correlation with patient age, assay sensitivity, threshold value, and mammographic screening. J Clin Path. 2000;53:688–96. doi: 10.1136/jcp.53.9.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elledge RM, Osborne CK. Oestrogen receptors and breast cancer. BMJ. 1997;314:1843–4. doi: 10.1136/bmj.314.7098.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]