Abstract
The Saccharomyces cerevisiae inositolsphingolipid phospholipase C (Isc1p), a homolog of mammalian neutral sphingomyelinases, hydrolyzes complex sphingolipids to produce ceramide in vitro. Epitope-tagged Isc1p associates with the mitochondria in the post-diauxic phase of yeast growth. In this report, the mitochondrial localization of Isc1p and its role in regulating sphingolipid metabolism were investigated. First, endogenous Isc1p activity was enriched in highly purified mitochondria, and western blots using highly purified mitochondrial membrane fractions demonstrated that epitope-tagged Isc1p localized to the outer mitochondrial membrane as an integral membrane protein. Next, LC/MS was employed to determine the sphingolipid composition of highly purified mitochondria which were found to be significantly enriched in α-hydroxylated phytoceramides (21.7 fold) relative to the whole cell. Mitochondria, on the other hand, were significantly depleted in sphingoid bases. Compared to the parental strain, mitochondria from isc1⊿ in the post-diauxic phase showed drastic reduction in the levels of α-hydroxylated phytoceramide (93.1 % loss compared to WT mitochondria with only 2.58 fold enrichment in mitochondria compared to whole cell). Functionally, isc1⊿ showed a higher rate of respiratory-deficient cells after incubation at high temperature and was more sensitive to hydrogen peroxide and ethidium bromide, indicating that isc1⊿ exhibits defects related to mitochondrial function. These results suggest that Isc1p generates ceramide in mitochondria, and the generated ceramide contributes to the normal function of mitochondria. This study provides a first insight into the specific composition of ceramides in mitochondria.
Keywords: Mitochondria, Sphingolipid, Sphingomyelinase, Lipidomics, ISC1, Yeast
1. Introduction
Sphingolipids play crucial roles as signaling molecules in eukaryotic cells, regulating such processes as stress responses, cell growth, differentiation, cell cycle arrest, endocytosis, and apoptosis [1-7]. Additionally, complex sphingolipids serve structural functions in membranes, and they may also undergo dynamic clustering with sterols to form lipid microdomains or ‘rafts’ within the membrane bilayer. This domain structure has been proposed to play a role as platforms for signal transduction and/or protein sorting [8].
Most, if not all, organelles contain sphingolipids as well as several sphingolipid-metabolizing enzymes [9-11]. Moreover, the hydrophobicity of bioactive sphingolipids restrains them to their specific compartments (unless transported by specific mechanisms). These considerations led to the proposal that sphingolipid function is compartment-specific [1]. In this context, many studies in eukaryotes have recently started suggesting important roles for mitochondrial sphingolipids [11-13].
Ceramide in mitochondria has been shown to regulate apoptosis in mammalian cells. For example, when bacterial sphingomyelinase was targeted to mitochondria, it induced mitochondrial ceramide generation and apoptosis [13], and mitochondrial ceramide has been implicated in the activation/translocation of the pro-apoptotic Bax [14]. Moreover, many studies have reported that ceramide has biophysical effects on mitochondrial membranes, by increasing the permeability of the outer and inner mitochondrial membranes [15], suppressing respiratory chain activity [16], or forming channels [17]. Therefore, the role of ceramide in mitochondria is attracting wide interests in terms of basic biology and medical application. Other studies have suggested mitochondrial localization of some enzymes of sphingolipid metabolism, including ceramide and dihydroceramide synthase [12, 18], ceramidase [19], glycosyltransferase [20], human sialidase [21], and sphingosine kinase [22].
Saccharomyces cerevisiae has proven to be an invaluable model for studying sphingolipid biosynthesis, in part because of its simplicity in sphingolipid metabolism relative to mammalian systems [3, 23]. Interestingly, the S. cerevisiae Isc1p, the homolog of mammalian neutral sphingomyelinases, was recently shown to associate with mitochondria in the post-diauxic phase of yeast growth [24]. Isc1p produces ceramide from the complex sphingolipids inositol phosphorylceramide (IPC), mannosylinositol phosphorylceramide (MIPC), and mannosyldiinositol phosphorylceramide (M(IP)2C) [25-28]. Isc1p was also shown to have a role in utilization of non-fermentable carbon sources, as an isc1⊿ strain grew very slowly in the post-diauxic phase or when grown in media containing non-fermentable carbon sources [29]. Utilization of non-fermentable carbon sources requires intact mitochondrial function, thus suggesting a role for Isc1p in mitochondrial function.
In this study we sought to elucidate the submitochondrial localization of Isc1p, its role in mitochondrial sphingolipid metabolism, and mitochondria-related phenotypes of isc1⊿. These studies have implications for compartment-specific sphingolipid metabolism and function in vivo.
2. Materials and Methods
2.1. Yeast strains and media
JK93α WT (MATα leu2−3, 112 ura3−52 rme1 trp1 his4) and JK93α isc1⊿ (MATα leu2−3, 112 ura3−52 rme1 trp1 his4 ISC1::G418) [25] were used in this study. Standard yeast culture medium was prepared using Difco YPD broth. Plates were prepared using Difco YPD agar and stored at 4°C. YPGE plates contained 1% yeast extract, 2% bactopeptone, 2% ethanol, 2% glycerol and 2% agar. Yeast cells were cultured at 30°C with shaking unless otherwise indicated.
2.2. Isolation of purified mitochondria
Isolation of purified mitochondria was basically performed essentially as described [30-32]. Briefly, yeast cells were incubated to saturation at 30°C. Six liter flask containing approximately 1.25 L YPD were inoculated with an overnight culture to OD600=0.1 and incubated at 30°C for 24 h with vigorous aeration. Yeast cells were centrifuged at 2,000 g for 5 min, washed with water, resuspended in 100 mM Tris-SO4, pH 9.4, 10 mM DTT (2 ml/g wet cell), incubated at 30°C for 15 min, centrifuged at 2,000 g for 5 min, washed with SP buffer (1.2 M sorbitol, 20 mM K-P pH 7.4), dissolved in SP buffer containing zymolyase 100T (7 ml/g cells, 1.5 mg/g cells), incubated at 30°C for 60 min, centrifuged at 2,500 g for 5 min, washed with SP buffer (7 ml/g wet cell) twice, resuspended in homogenization buffer (0.6 M sorbitol, 10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) (6.5 ml/g cells), homogenized in a 40 ml dounce homogenizer coated with Teflon using 25 strokes, diluted two fold with homogenizing buffer, centrifuged for 5 min at 1,500 g to remove cell nuclei and debris, and centrifuged again at 2,500 g for 5 min. The supernatant was centrifuged at 12,000 g for 15 min. The supernatant was saved as the post-mitochondrial fraction and TCA-precipitated for further analysis. The pellet was resuspended in 30 ml SM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) and centrifuged at 2,500 g for 5 min. The supernatant was then centrifuged at 12,000 g for 15 min, and the pellet was resuspended in 10 ml SM buffer as crude mitochondria. For purification of mitochondria, the amount of mitochondrial protein was measured by OD280, adjusted to 5 mg/ml protein in SM buffer, homogenized with 10 strokes in a dounce homogenizer coated with Teflon, and subsequently loaded onto a four step sucrose gradient [1.5 ml of 60%, 4 ml of 32%, 1.5 ml of 23%, 1.5 ml of 15% of sucrose in 1 mM EDTA, 10 mM MOPS-KOH (pH 7.2)], and centrifuged at 134,000g for 1 h with no braking. The 32/60% interface was then recovered with a Pasteur pipette and washed with 2 vol of SM buffer at 12,000 g for 15 min. The mitochondrial fraction was again homogenized with 10 strokes in a dounce homogenizer coated with Teflon and then applied to a four step sucrose gradient as described above. The 32/60% interface was recovered, resuspended in 2 volumes of SM buffer, and centrifuged at 12,000 g for 15 min. The final pellet was resuspended in SM buffer, divided into small aliquots, and frozen at −80°C.
2.3. LC/MS measurements
Purified mitochondria or yeast cells in the post-diauxic phase were snap-frozen in a methanol-dry ice bath and stored at −80°C. ESI/MS/MS analysis of endogenous phyto- and dihydrosphingosine bases, their phosphates, α-hydroxylated phytoceramide, and non-hydroxylated phyto- and dihydroceramide species were performed on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer, operating in a Multiple Reaction Monitoring positive ionization mode, using modified version [28, 34]. Briefly, biological samples were fortified with internal standards (ISs: C17 base D-erythro-sphingosine, C17 sphingosine-1-phosphate, N-palmitoyl-D-erythro-C13 sphingosine, heptadecanoyl-D-erythro-sphingosine and C6-Phyto-ceramide), then extracted with 70% isopropanol:ethylacetate:pyridine:25% ammonia (60:20:2:0.5 by volume) solvent system. After evaporation and reconstitution in 150 μl of methanol, samples were injected on the HP1100/TSQ 7000 LC/MS system and gradient eluted from the BDS Hypersil C8, 150 × 3.2 mm, 3 μm particle size column, with 1.0 mM methanolic ammonium formate / 2 mM aqueous ammonium formate mobile phase system. Peaks corresponding to the target analytes and ISs were collected and processed using the Xcalibur software system. Quantitative analysis was based on the calibration curves generated by spiking an artificial matrix with the known amounts of the target analyte synthetic standards and an equal amount of IS. The target analyte/IS peak areas ratios were plotted against analyzed concentration. The target analyte/IS peak area ratios from the samples were similarly normalized to their respective ISs and compared to the calibration curves, using a linear regression model.
2.4. Submitochondrial localization of Flag-Isc1p
Submitochondrial localization of Flag-Isc1p was determined as previously described [35]. Briefly, 200 μg of highly purified mitochondria from isc1⊿+pYES/FLAG-ISC1 were pelleted at 10,000 g for 15 min and suspended in 40 μl SM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS-KOH, pH 7.2). Samples were split into two equal portions. One hundred and eighty microliters of SM buffer or EM buffer (1 mM EDTA, 10 mM MOPS-KOH, pH 7.2) was added, pipetted up and down 10 times to facilitate the rupture of the outer membrane from the swollen mitochondria, and incubated for 15 min on ice. Samples were split into two equal portions and treated with or without 50 μg/ml proteinase K for 30 min on ice. Phenylmethylsulfonyl fluoride was added to the solution (final concentration 2 mM). The sample was then centrifuged at 12,000×g for 5 min, washed once and analyzed by western blotting using anti-Flag antibody.
2.5. Determination of association of proteins with membrane
Highly purified mitochondria, purified from isc1⊿+pYES/FLAG-ISC1, were treated without osmotic shock (250 mM sucrose, 10 mM HEPES, pH 7.2, 1 mM EDTA), with osmotic shock (10 mM HEPES, pH 7.2, 1 mM EDTA), with osmotic shock and high salt (0.1 M NaCl, 10 mM HEPES, pH 7.2, 1 mM EDTA), or with osmotic shock and alkali pH (0.1 M Na2CO3, pH 11.2, 10 mM HEPES, pH 7.2, 1 mM EDTA). They were then centrifuged at 100,000×g for 10 min, and the pellet and supernatant were analyzed by western blotting using antibody against Flag.
2.6. Statistical analysis
The amounts of sphingolipid species of mitochondria prepared from WT and isc1⊿ are presented as means ± standard error of the mean (SEM) of three lipid measurements from two independent cultures and mitochondrial purifications. The amounts of sphingolipid species of the whole cell are presented as means ± SEM of lipid measurements from three independent cultures. Statistical differences were evaluated by Student's unpaired t test.
2.7. Miscellaneous
Antibodies against Cox3p, Porin, Dpm1p, V-ATPase (100kDa subunit), and Pgk1p were obtained from Molecular Probes, and that against FLAG was from Sigma-Aldrich. The antibody against Mge1p was a kind gift from Dr. C. M. Koehler.
3. Results
3.1. Isc1p as an integral membrane protein localized to the outer membrane of mitochondria
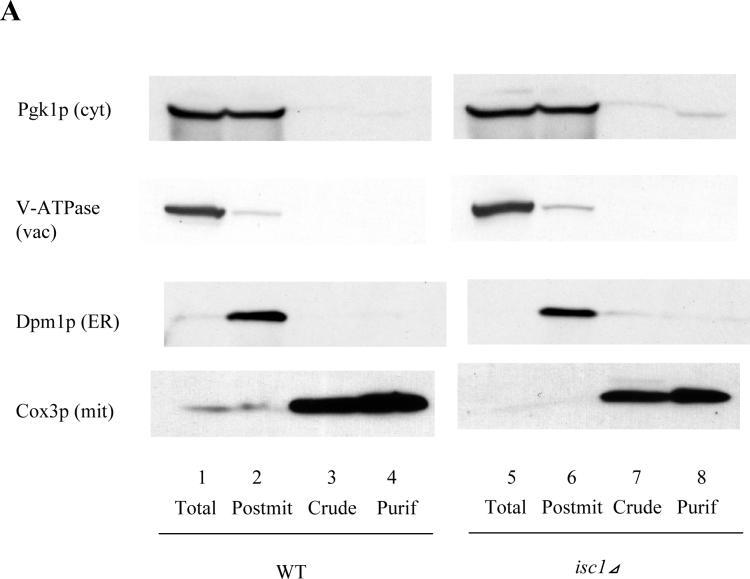
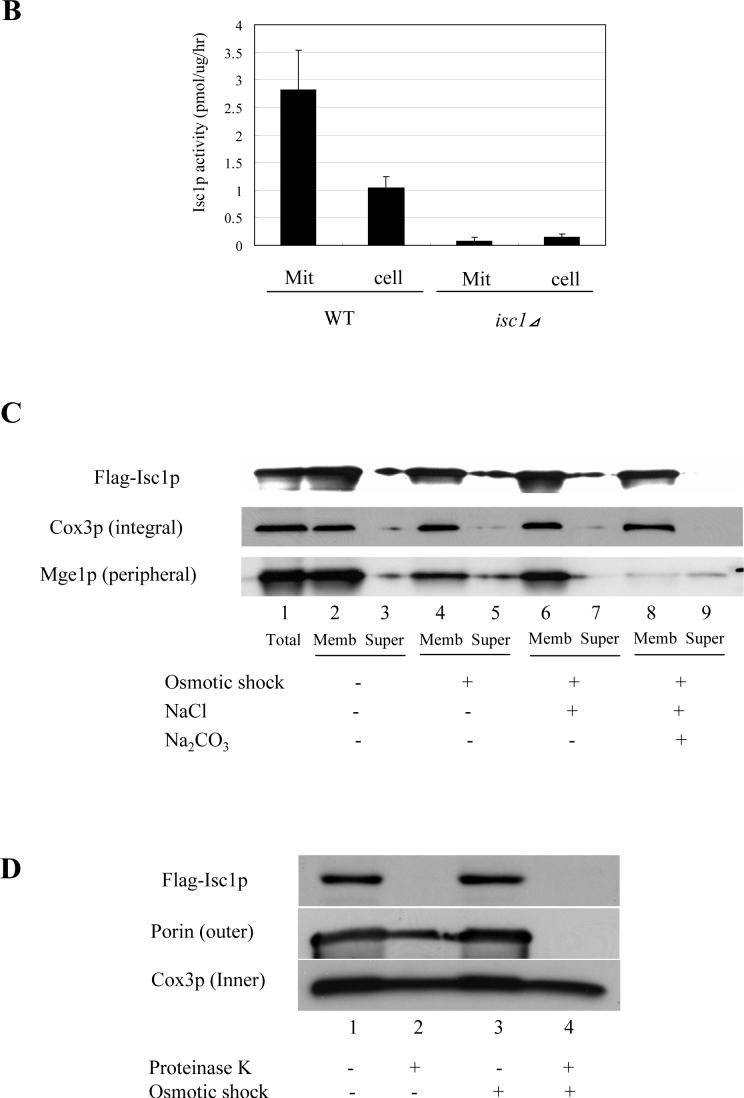
Since epitope-tagged Isc1p was previously shown to associate with mitochondria in the post-diauxic phase, it became important to determine the mitochondrial localization of endogenous Isc1p. WT and isc1⊿ cells were incubated to the post-diauxic phase, collected by centrifugation, and mitochondria were isolated to a high degree of purity using multi-step centrifugation in conjunction with a two-step sucrose gradient, which has been verified by mitochondrial proteome studies [30-33]. The purity of the mitochondria was confirmed by western blotting with an antibody against a cytosolic marker protein, Pgk1p, a vacuolar marker protein, V-ATPase, an endoplasmic reticulum (ER) marker protein, Dpm1p, and a mitochondrial marker protein, Cox3p. The results showed that the purified fraction was devoid of Pgk1p, V-ATPase and Dpm1p and highly enriched in Cox3p (Fig. 1A), indicating that mitochondria were purified to a high degree, consistent with previous reports [30-32]. Subsequent studies were performed using these highly purified mitochondria prepared from post-diauxic phase cells. In order to determine the localization of endogenous Isc1p, Isc1p enzymatic activity in the purified mitochondria was determined. Isc1p specific activity was highly concentrated in the mitochondrial fraction (2.8 pmol/μg/hr) when compared to the specific activity of the whole cell homogenate (1.0 pmol/μg/hr) (Fig. 1B). Moreover, cells and mitochondria from the isc1⊿ strain contained only background activity, demonstrating the Isc1p is the primary enzyme responsible for the detected activity in mitochondria. Taken together, these results demonstrate that Isc1p activity is indeed localized to mitochondria (Fig. 1B).
Fig. 1. Purification of mitochondria and submitochondrial fractionation of Flag-Isc1p.
(A) Validation of the purity of mitochondria. Forty micrograms of each sample (lanes 1,5; total cell lysate, lanes 2,6; post-mitochondrial fraction, lanes 3,7; crude mitochondria and lanes 4,8; purified mitochondria) from WT (lanes 1−4) and isc1⊿ (lanes 5−8) were subjected to western blotting against a cytosolic protein, Pgk1p, a vacuolar protein, V-ATPase, an ER membrane protein, Dpm1p, and a mitochondrial membrane protein, Cox3p. (B) Isc1p activity of purified mitochondria. Isc1p activity of purified mitochondria and the whole cell from WT and isc1⊿ was measured basically as previously described (24). (C) Membrane association of Flag-Isc1p. Mitochondria purified from isc1 ⊿+pYES/FLAG-ISC1 were treated without osmotic shock (lanes 2, 3; 250 mM sucrose, 10 mM HEPES, pH 7.2, 1 mM EDTA), with osmotic shock (lanes 4, 5; 10 mM HEPES, pH 7.2, 1 mM EDTA), with osmotic shock and high salt (lanes 6, 7; 0.1 M NaCl, 10 mM HEPES, pH 7.2, 1 mM EDTA), or with osmotic shock and alkali pH (lanes 8, 9; 0.1 M Na2CO3, pH 11.2, 10 mM HEPES, pH 7.2, 1 mM EDTA), centrifuged at 100,000 g for 10 min, and total (lane 1), membrane (lanes 2, 4, 6, 8), and supernatant (lanes 3, 5, 7, 9) were analyzed by western blotting using an antibody against Flag. (D) Submitochondrial localization of Flag-Isc1p. Mitochondria purified from isc1⊿+pYES/FLAG-ISC1 were treated without osmotic shock (lanes 1, 2; 250 mM sucrose, 10 mM HEPES, pH 7.2, 1 mM EDTA) or with osmotic shock (lanes 3, 4; 10 mM HEPES, pH 7.2, 1 mM EDTA) for 10 min on ice and with (lanes 2, 4) or without (lanes 1, 3) final 50 μg/ml proteinase K for 30 min on ice. 2 mM phenylmethylsulfonyl fluoride was added, mitochondria were centrifuged at 12,000 g for 5 min, washed once, and pelles were applied to western analysis with antibody against Flag, porin, or Cox3p.
Next it became important to determine if Isc1p is loosely associated with mitochondria or if it is an integral mitochondrial membrane protein. Mitochondria were treated with osmotic shock (to lyze the mitochondrial outer membrane) and then treated with either high salt or alkali pH extraction, centrifuged at 100,000 g for 10 min, and pellets were analyzed by western blotting. N-terminally tagged Flag-Isc1p [25] was not dissociated from the membrane with osmotic shock (Fig. 1C lanes 4 and 5), high salt (Fig. 1C lanes 6 and 7) or alkali pH (Fig. 1C lanes 8 and 9). Cox3p, a known integral membrane protein behaved similarly whereas Mge1p, a peripheral mitochondrial protein became significantly dissociated from membranes in response to treatment with alkaline pH. Taken together, these results indicate that Isc1p is a bona fide membrane protein.
Next, the submitochondrial localization of Isc1p was determined using protease protection assays. Proteinase K was added to purified mitochondria with or without osmotic shock, and the degradation of several marker proteins was examined by western blotting. Using this assay without osmotic shock, exogenously added proteinase K can only access and degrade proteins exposed from the outer mitochondrial membrane. On the other hand, with osmotic shock, proteinase K causes degradation of proteins contained in the outer membrane, in the intermembrane space, or exposed out of the inner membrane since osmotic shock causes rupture of the outer mitochondrial membrane while leaving the inner mitochondrial membrane intact. A previously characterized inner membrane protein, Cox3p, was not degraded even after osmotic shock (Fig. 1D, lane 4), demonstrating that under these conditions, no protease cleavage sites are exposed out of the inner membrane. On the other hand, porin, a marker protein for outer mitochondrial membrane proteins, was degraded only after osmotic shock (Fig. 1D, lane 4), suggesting that it is inserted in the outer membrane so that it is degraded only after rupture of the outer membrane. To the contrary, Flag-Isc1p was efficiently degraded even without osmotic shock (Fig. 1D, lane 2), indicating that Flag-Isc1p localized to the outer membrane of mitochondria with exposed protease cleavage sites, or, alternatively, only N-terminally tagged FLAG is exposed from the outer membrane and Isc1p is localized inside the outer membrane. From all the above data, it could be concluded that Flag-Isc1p localizes to the outer membrane of mitochondria and is integrated into the outer mitochondrial membrane.
3.2. Mitochondria are enriched in very long chain phytoceramides
In previous reports, we showed that Isc1p degrades complex sphingolipids such as IPC, MIPC and M(IP)2C to form ceramides in vitro [25-27] and that Isc1p is activated by phosphatidylglycerol and cardiolipin [29], two phospholipids that reside in mitochondria [36,37]. These results, together with the mitochondrial localization of Isc1p, raised the possibility that Isc1p may be responsible for generation of ceramides in mitochondria in vivo. To address this issue, we first determined the sphingolipid composition of mitochondria and compared it to that of whole cells. WT cells were incubated to the post-diauxic phase, and the mitochondria were isolated to a high degree of purity using a two-step sucrose gradient as described above. The sphingolipid profile of the whole cell of WT and the mitochondria purified from WT was determined using LC-MS [34]. The masses of sphingolipids were calculated based on the phospholipid content.
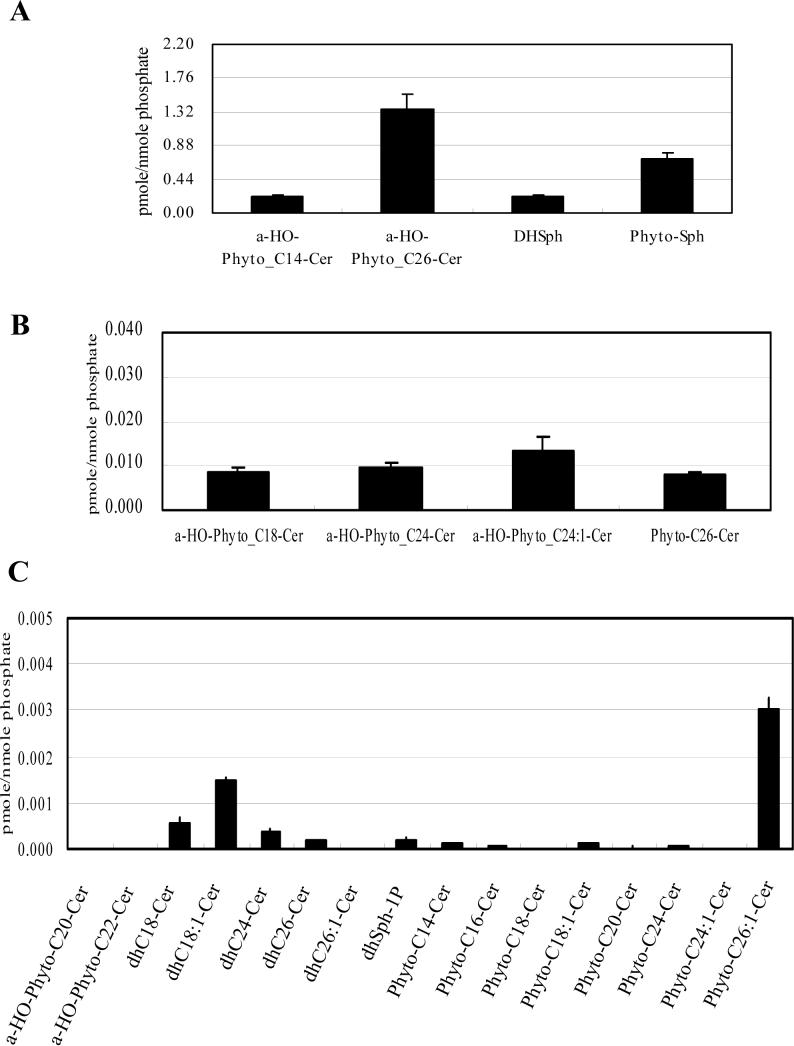
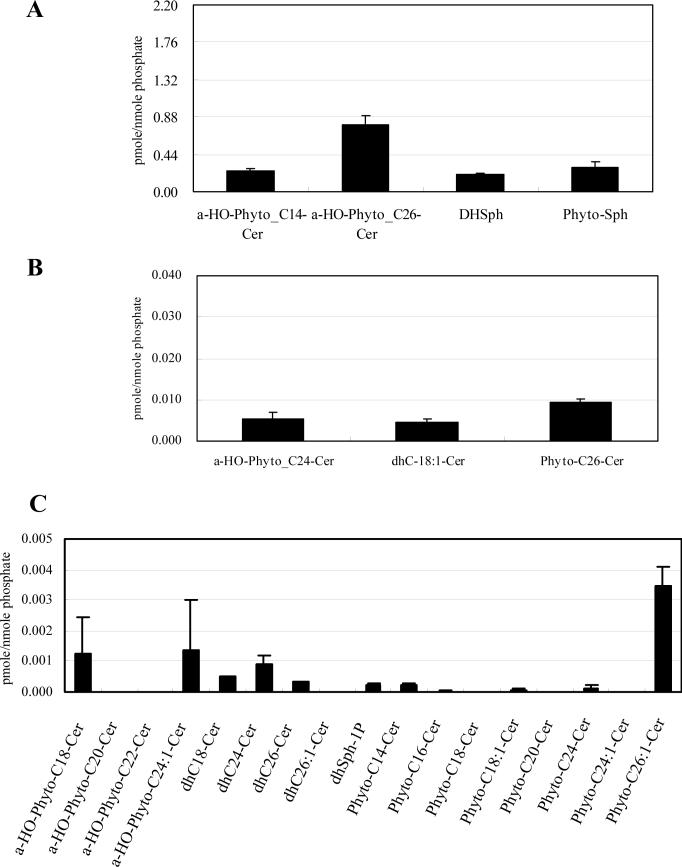
In whole cells, α-hydroxylated C26-phytoceramide, phytosphingosine, dihydrosphingosine, and α-hydroxylated C14-phytoceramide constituted the major species (Fig. 2A), α-hydroxylated C24:1-, C24- and C18-phytoceramides and non-hydroxylated C26-phytoceramides were next in abundance (Fig. 2B), and α-hydroxylated C20- and C22-phytoceramides and most of the non-hydroxylated dihydroceramides and phytoceramides showed significantly lower levels (Fig. 2C).
Fig. 2. Sphingolipid profile of WT cells.
WT cells were incubated to the post-diauxic phase. Sphingolipids were analyzed by LC-MS and normalized by organic phosphates as described in Materials and Methods. (A) Species which are more than 0.040 pmol/nmol organic phosphate, (B) species which are more than 0.0050 pmol/nmol organic phosphate but less than 0.040 pmol/nmol organic phophate, and (C) species which are less than 0.0050 pmol/nmol organic phosphate of the whole cell of WT. The results are mean values +/− SEM of lipid measurements from three independent cultures.
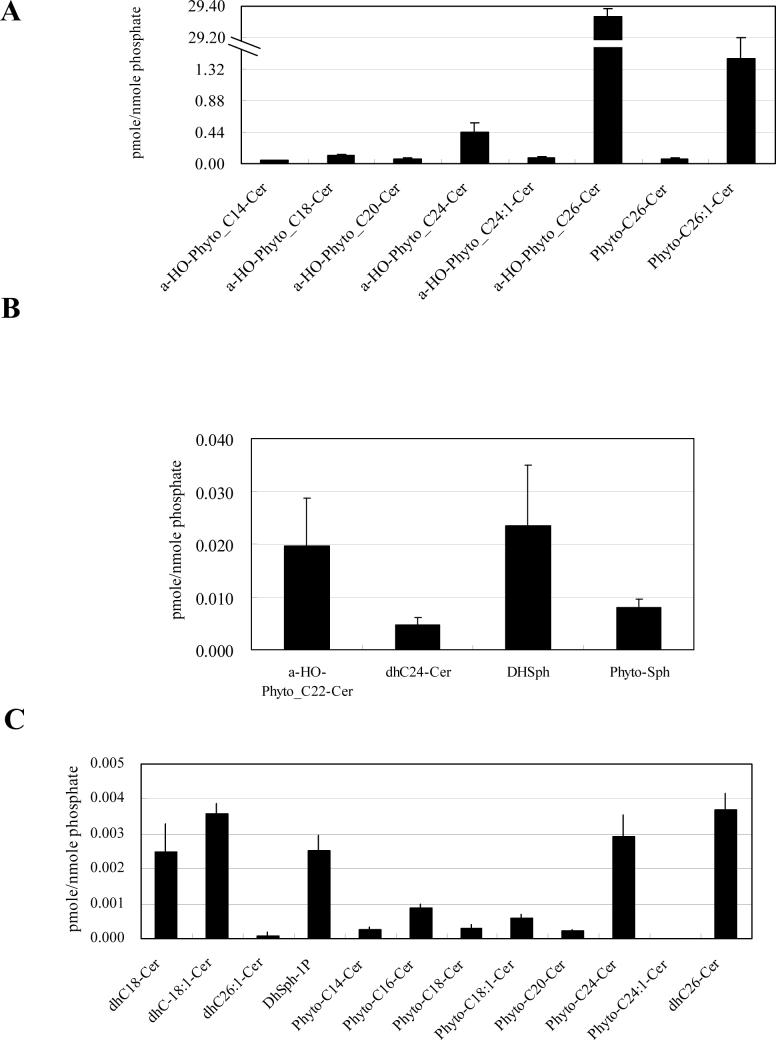
In contrast, the results revealed, for the first time, that mitochondria contain predominantly α-hydroxylated C26-phytoceramide (Fig. 3A) with significantly lower levels of other ceramides or sphingoid bases (Fig. 3B and 3C). These results were expressed relative to mitochondrial total phospholipids, and therefore, they represent the relative surface concentration of these sphingolipids in membranes. It should be noted that even for the highest ceramide, the ratio to phospholipids was less than 3%, thus demonstrating that phospholipids are indeed in excess and that mitochondrial ceramides are minor species.
Fig. 3. Sphingolipid profile of WT mitochondria.
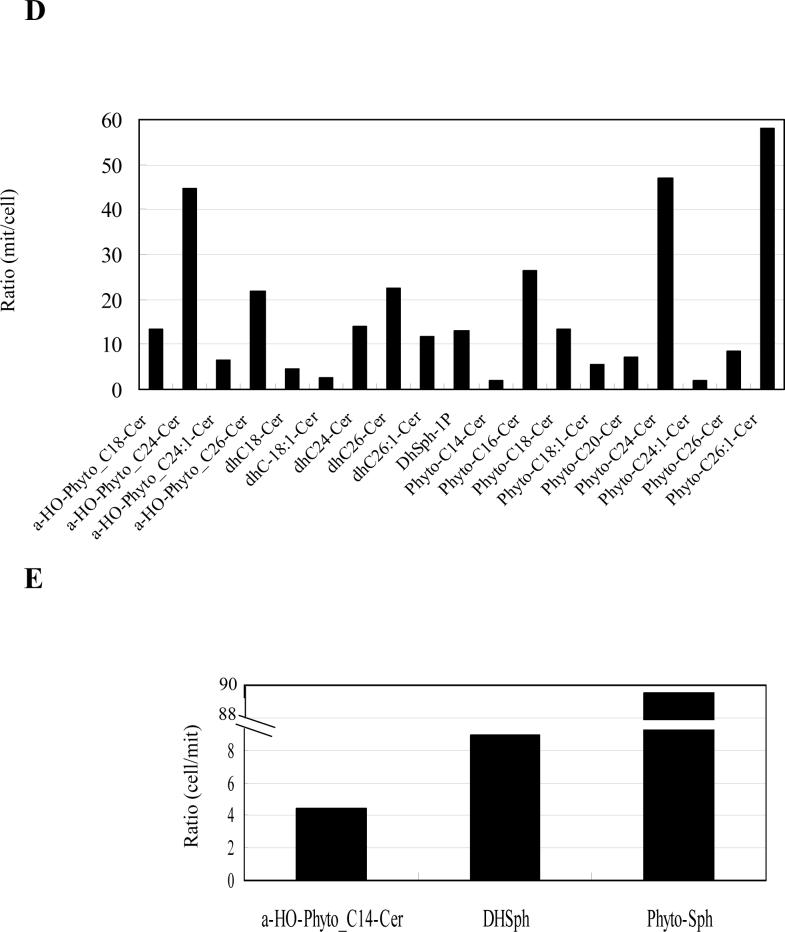
WT cells were incubated to the post-diauxic phase, and mitochondria were purified as described in Materials and Methods. Sphingolipids were analyzed by LC-MS and normalized by organic phosphates as described in Materials and Methods. (A) Species which are more than 0.040 pmol/nmol organic phosphate, (B) species which are more than 0.0050 pmol/nmol organic phosphate but less than 0.040 pmol/nmol organic phophate, and (C) species which are less than 0.0050 pmol/nmol organic phosphate of the WT mitochondria. The results are mean values +/− SEM of three lipid measurements from two independent mitochondrial purifications. Mitochondrial sphingolipids were divided by the amount of sphingolipids in the whole cell analyzed in Fig. 2. (D) Ratio of mitochondrial sphingolipids to the sphingolipids of the whole cell. Sphingolipids that were increased in mitochondria are shown. (E) Ratio of the sphingolipids in the whole cell to mitochondrial sphingolipids. Sphingolipids that were decreased in mitochondria are shown.
In order to determine the relative enrichment of mitochondrial ceramides to that of whole cells, we calculated the degree of ‘surface enrichment’ of these sphingolipids in mitochondria to whole cells by dividing the ceramide/phospholipid ratio in mitochondria over that in whole cells. This calculated ratio of mitochondrial/whole cell for α-hydroxylated C26-phytoceramides was 21.7 fold, and this enrichment in mitochondria was also observed for other ceramide species (Fig. 3D), indicating that mitochondria were highly enriched with ceramide species mainly composed of α-hydroxylated phytoceramides. This result also suggests that the major source of α-hydroxylated phytoceramides of the whole cell is derived from the mitochondria. Non-hydroxylated ceramides also showed increases in the mitochondria relative to the whole cells (Fig. 3D). Notably, the levels of the non-hydroxylated phytoceramides and dihydroceramides were significantly lower than those of α-hydroxylated phytoceramides both in whole cells and mitochondria (Fig. 2 and Fig. 3).
Reciprocally, the levels of phytosphingosine, the predominant ‘simple’ sphingolipid of yeast, were significantly decreased in mitochondria relative to whole cells (Fig. 3E, note inverted y-axis compared to 3D).
Taken together, the results demonstrate that in S. cerevisiae, mitochondria show selective enrichment in α-hydroxylated phytoceramides and selective depletion of sphingoid bases. These results were basically similar when sphingolipid levels were normalized by protein amount (data not shown), indicating that these results are not due to selective difference in total phospholipid levels between the whole cell and mitochondria.
3.3. Mitochondria of isc1⊿ show an altered sphingolipid profile
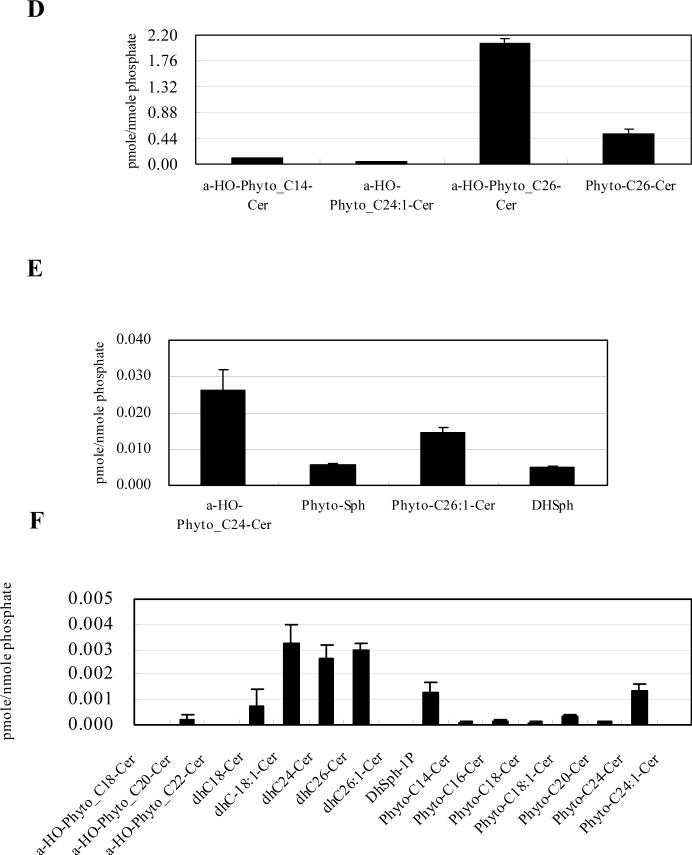
Next, the sphingolipid profile of mitochondria prepared from isc1⊿ was determined and compared to that of WT mitochondria in order to determine the contribution of Isc1p to mitochondrial sphingolipids. The major sphingolipids detected in whole cells of the isc1⊿ strain were α-hydroxylated C26-phytoceramide, phytosphingosine, α-hydroxylated C14-phytoceramide and dihydrosphingosine (Fig. 4A) followed by non-hydroxylated C26-phytoceramide, α-hydroxylated C24-phytoceramide and non-hydroxylated C18:1-dihydroceramide (Fig. 4B), with other ceramides showing very low levels (Fig. 4C).
Fig. 4. Sphingolipid profile of isc1⊿.
isc1⊿ cells were incubated to the post-diauxic phase, and collected for whole cell analyses or for mitochondrial purification as described in Materials and Methods. Sphingolipids were analyzed by LC-MS as described in Materials and Methods, and normalized by organic phosphates. (A) Species which are more than 0.040 pmol/nmol organic phosphate, (B) species which are more than 0.0050 pmol/nmol organic phosphate but less than 0.040 pmol/nmol organic phophate, (C) species which are less than 0.0050 pmol/nmol organic phosphate of isc1⊿ whole cell. (D) Species which are more than 0.040 pmol/nmol organic phosphate, (E) species which are more than 0.0050 pmol/nmol organic phosphate but less than 0.040 pmol/nmol organic phophate, and (F) species which are less than 0.0050 pmol/nmol organic phosphate of isc1⊿ mitochondria. The results are mean values +/− SEM of lipid measurements from three independent cultures for the whole cell analysis, or, three lipid measurements from two independent mitochondrial purifications for mitochondrial analysis.
The predominant species of sphingolipids in mitochondria from isc1 ⊿ were α-hydroxylated C26-phytoceramide, non-hydroxylated C26-phytoceramide, α-hydroxylated C14-phytoceramide and α-hydroxylated C24:1-phytoceramide (Fig. 4D), followed by α-hydroxylated C24-phytoceramide, non-hydroxylated C26:1-phytoceramide, phytosphingosine and dihydrosphingosine (Fig. 4E), with other sphingolipids being minor species (Fig. 4F).
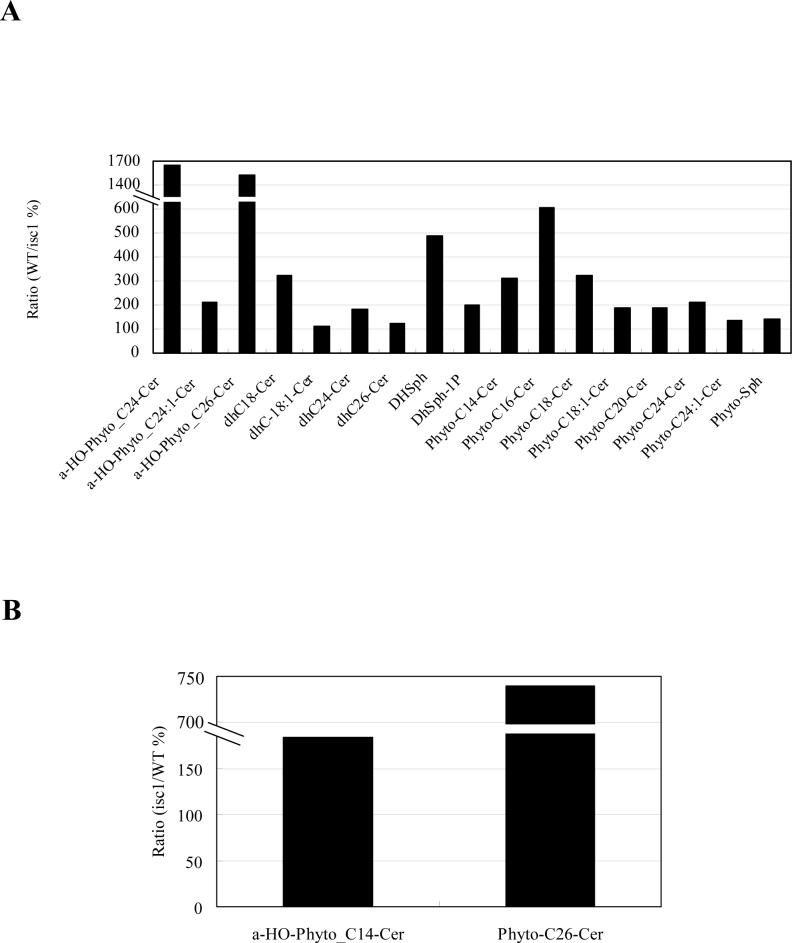
Many of these species showed significant reduction in their enrichment in mitochondria. Most notably, the content of α-hydroxylated C26-phytoceramide in isc1⊿ mitochondria was only 6.94% of that of WT mitochondria (Fig. 5A), and this decrease was significantly rescued by expression of FLAG-ISC1 (Fig. 5C). As a consequence, the enrichment of α-hydroxylated C26-phytoceramide in mitochondria was decreased to 2.58 fold in isc1⊿ (Fig. 4A and 4D) relative to the 21.7 fold in WT (Fig. 3D). This decrease in isc1⊿ mitochondria was also observed for other ceramides (Fig. 5A).
Fig. 5. Comparison of mitochondrial sphingolipids of isc1⊿ with those of WT.
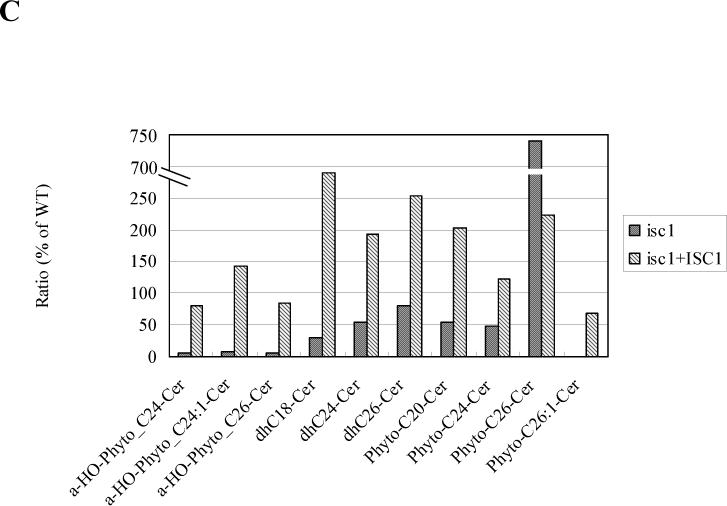
The relative amounts of individual mitochondrial sphingolipids of isc1⊿ and isc1⊿+pYES/FLAG-ISC1 were compared with those of WT. (A) Ratio of the sphingolipids in WT relative to those in isc1⊿ . Sphingolipids that were decreased in isc1⊿ are shown. (B) Ratio of the sphingolipids in isc1⊿ relative to those in WT. Sphingolipids that were increased in isc1⊿ are shown. (C) Ratio of the sphingolipids in isc1⊿ and isc1⊿+pYES/FLAG-ISC1 relative to those in WT.
These results suggest that Isc1p functions in cells to generate mitochondrial ceramides mainly composed of α-hydroxylated phyotceramides. Reciprocally, α-hydroxylated C14-phytoceramide and non-hydroxylated C26-phytoceramide were increased in isc1⊿ mitochondria (Fig. 5B), suggesting a complementary pathway to overcome the decrease of other ceramide species. These lipid changes in isc1⊿ mitochondria were partially suppressed by the expression of FLAG-ISC1 (Fig. 5C), although some of the lipids were not recovered to 100%, probably because FLAG-ISC1 was expressed under the GAL1 promoter. Nontheless, this result indicates that the lipid changes observed in isc1⊿ mitochondria relative to WT mitochondria are not derived from artifact of mitochondrial purification. Phospholipid content per protein in WT mitochondria was 34.9+/−3.14 (nmol/mg), while that in isc1⊿ mitochondria is 36.7+/−0.96 (nmol/mg), indicating that the mass profile changes induced by the deletion of ISC1 are not due to phospholipid changes in the mitochondria of isc1⊿. These results demonstrate that the isc1⊿ has an altered mitochondrial sphingolipid profile, and suggest that Isc1p produces selectively α-hydroxylated phytoceramides in mitochondria in vivo.
3.4. Disruption of ISC1 induces mitochondrial-associated defects
The alteration of sphingolipid profile of mitochondria in vivo observed above suggested that Isc1p may influence mitochondrial related functions. In order to address this hypothesis, we examined the formation of petite colonies in isc1⊿. In S. cerevisiae, mitochondrial defects often manifest by growing as respiratory deficient, “petite” colonies on fermentable carbon sources. In addition, cells with compromised mitochondria form large numbers of petite colonies after prolonged culture at elevated temperature when grown on either fermentable or non-fermentable carbon sources [38,39]. To determine whether isc1⊿ would form petites under these conditions, the isc1⊿ strain was grown at normal (30°C) and high temperature (39°C), and the fraction of petite colonies was determined by counting the ratio of colonies which have smaller size than normal and are unable to grow on non-fermentable carbon source plates (YPGE). The results showed that the isc1⊿ strain exhibited a significantly increased ratio of petite colonies at normal temperature (p<0.01, Fig. 6A) and this phenotype was significantly exacerbated by growth at high temperature (p<0.12, Fig. 6A).
Fig. 6. isc1⊿ shows mitochondrial defects.
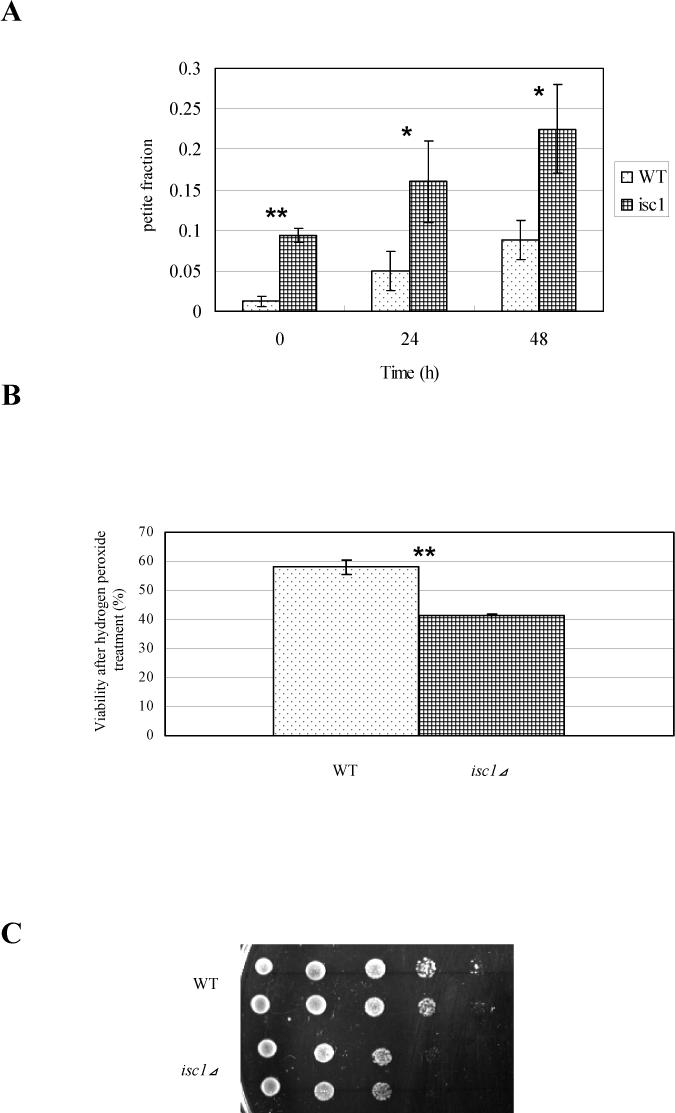
(A) Petite ratio of isc1⊿ . WT and isc1⊿ cells were inoculated at OD600 of 0.1 in YPD medium, incubated at 39°C for the indicated times, and plated onto YPD plates. Small colonies that grew on YPD plates were replica plated onto YPGE plates to confirm the inability to grow on non-fermentable carbon medium, and the numbers of the colonies which grew on YPD but not on YPGE plates were counted. Statistical differences were evaluated by Student's unpaired t test and are indicated by asterisks (**p<0.01, *p<0.12). (B) Hydrogen peroxide sensitivity of isc1⊿. WT and isc1⊿ were treated with 1 mM hydrogen peroxide for 45 min, plated onto YPD and the formed colonies were counted. Statistical difference was evaluated by Student's unpaired t test and is indicated by asterisks (**p<0.01). (C) Ethidium bromide sensitivity of isc1⊿. WT and isc1⊿ cells were treated with 50 μM ethidium bromide, and 5 μl of the suspension was spotted onto a YPD plate with serial dilutions at 10 fold.
Yeast cells with compromised mitochondria have also been shown to be sensitive to oxidative stress such as hydrogen peroxide [40]. To test whether the isc1⊿ strain was sensitive to hydrogen peroxide, yeast in log phase growth were treated with 1 mM hydrogen peroxide for 45 min, and the number of colonies was measured. Figure 6B demonstrates that the number of colonies of WT cells treated with hydrogen peroxide was 58.0% of that of non-treated cells whereas the number of colonies of isc1⊿ cells treated with hydrogen peroxide was 41.2% of that of non-treated cells, indicating that isc1⊿ is significantly sensitive to hydrogen peroxide relative to WT (p<0.01).
Ethidium bromide has been reported to specifically damage mitochondrial DNA, and mutants that have dysfunctional mitochondria show altered sensitivity to this substance [41,42]. To elucidate if isc1⊿ is sensitive to ethidium bromide, WT and isc1⊿ cells were treated with 50 μM ethidium bromide and spotted onto a YPD plate. The results showed that isc1⊿ was more sensitive to ethidium bromide than WT (Fig. 6C). Together, these results indicate that isc1⊿ has defective mitochondrial phenotypes and reveal a role of sphingolipid in mitochondrial function in vivo.
4. Discussion
Data in this report reveal that Isc1p of Saccharomyces cerevisiae localizes to the outer membrane of mitochondria as an integral membrane protein, and its activity is enriched in purified mitochondria. Moreover, this study is the first to measure α-hydroxylated ceramide species and to show that mitochondria are highly enriched in very long chain α-hydroxylated phytoceramides. Importantly isc1⊿ showed an altered mitochondrial sphingolipid profile, indicating that Isc1p regulates sphingolipid metabolism of mitochondria, especially the formation of α-hydroxylated very long chain phytoceramides. Finally, isc1⊿ displayed defects in mitochondrial functions, suggesting, for the first time, a role for mitochondrial sphingolipid metabolism in mitochondrial metabolic function in vivo.
Recent advances in methods for mitochondrial purification as well as newly developed lipidomic strategies have enabled a more comprehensive and precise assessment of simple sphingolipids in mitochondria in Saccharomyces cerevisiae. Previous data indicated that mitochondria contain 7.4% of the total complex sphingolipid content, with an additional 30% in plasma membrane [10]. Another report demonstrated that mitochondria contain 4.6 nmol of IPC/mg protein, 7.7 nmol of MIPC/mg protein and 5.6 nmol of M(IP)2C/mg protein, while plasma membrane contains 34.9 nmol of IPC/mg protein, 69.9 nmol of MIPC/mg protein and 120.5 nmol of M(IP)2C/mg protein [9]. It is important to note, however, that the mitochondrial fractions used for these studies were not derived from density gradient centrifugation, and thus, it is difficult to know the degree of contamination of these preparations with microsomal membranes. Moreover, these previous reports studied only complex sphingolipids and did not address ceramides and sphingoid bases, the subjects of this study.
With this current analysis, it was found that mitochondria from WT cells are highly enriched, and specifically, with very long chain α-hydroxylated phytoceramides. This unexpected result raises a number of questions as to the metabolism of very long chain sphingolipids in yeast and their function. Very little is known about the regulation of incorporation of very long chain fatty acids into sphingolipids or the subcellular sites of migration of these lipids. Both Lag1p and Lac1p ceramide synthases, thought to reside in the ER, are capable of introducing very long chain fatty acids into ceramides [43], which in turn are incorporated in IPC, MIPC, and M(IP)2C [44-47]. While the bulk of these complex sphingolipids eventually arrives at the plasma membrane, published results show that a significant component of these lipids may reside in the mitochondria [9]. Together with the current results that Isc1p is localized to the outer membrane of mitochondria (Fig. 1) and mitochondria purified from isc1 ⊿ exhibit a lower content of α-hydroxylated phytoceramides than that from WT (Fig. 5A), it is suggested that these complex sphingolipids become substrates for Isc1p either at the outer mitochondrial membrane or in a compartment that comes in contact with the outer membrane (e.g. mitochondrial associated membranes or others). Further studies are required to determine where and how Isc1p interacts with yeast complex sphingolipids. Moreover, this study did not address the issue of topology of Isc1p, and therefore, it is not known whether its active site faces into the mitochondria or out of the mitochondria. This proposal that sphingolipids are generated at the outer membrane of mitochondria is also in agreement with the previous report [48] that detected threefold enrichment of ceramide in the outer membrane relative to the inner membrane of rat liver mitochondria. Considering that Isc1p is activated by cardiolipin and phosphatidylglycerol [26, 29] and that these lipids are synthesized in the inner membrane of mitochondria [49-51], Isc1p may be activated at the membrane contact site of outer and inner membrane [52,53]. Alternatively, up to 36.6% of cardiolipin has been detected in the outer membrane of mitochondria [54,55] and this could then activate Isc1p. Further investigation is required for elucidation of pathways of subcellular transport and metabolism of complex sphingolipids.
The results also showed that mitochondria are relatively depleted in sphingoid bases, and this is consistent with the known ER sites for de novo sphingolipid synthesis [56] or ceramide degradation in the ER and Golgi [57,58].
The current results as well as previous results [29] implicating Isc1p in regulation of metabolism of non-fermentable carbon sources suggest a role for either the precursor sphingolipids and/or the generated α-hydroxylated phytoceramides in regulating metabolic functions of mitochondria. These could include signaling functions of the ceramides and/or structural functions. The latter are consistent with the recent suggestion that many mitochondrial proteins reside in ‘rafts’ [59]. Moreover, genome-wide analysis revealed that most of ergosterol synthetic mutants showed defective mitochondrial morphology [60]. Since, ergosterol has been reported to form rafts together with sphingolipids [61], the results in this study raise the possibility of sphingolipid/sterol structures/raft in the function of mitochondria.
The specific ceramide generated by the action of Isc1p could also play regulatory functions. In mammalian cells, ceramide, exogenously added or generated endogenously by overexpression of mitochondrially-targeted sphingomyelinase, has been shown to induce/regulate Bax translocation and to be involved in mitochondrial dysfunction, such as increases in the permeability of the outer membrane and release of cytochrome c [13, 15, 17, 62]. However, whether endogenous ceramides are generated in eukaryotic mitochondria and exert effects on mitochondria in vivo has not been well evaluated and has been under significant discussion [63]. The results in this study show, for the first time, that native alteration in sphingolipid metabolism in mitochondria causes dysfunctions of mitochondria in vivo, suggesting an intrinsic role of some specific ceramide in regulating mitochondrial functions. However, further studies are required for elucidation of sphingolipid-mediated functions in yeast.
In conclusion, we have determined the submitochondrial localization of Isc1p and its effect on mitochondrial sphingolipid metabolism, and elucidated that isc1⊿ exhibits mitochondria-related phenotypes. The findings contribute key knowledge toward complete understanding of the metabolism of mitochondrial sphingolipids and their roles in mitochondrial function in eukaryotic cells.
Acknowledgements
We thank Dr. Tatyana I. Gudz, Dr. Hiroko Hama, Dr. Kazuyuki Kitatani, Dr. Chiara Luberto, and Dr. Fernando Alvarez-Vasquez for helpful advice, Dr. Alicja Bielawska for lipid analyses and Dr. Carla M. Koehler for the antibody against Mge1p. Liquid-chromatography-MS lipid analyses were performed at the Lipidomics Core at the Medical University of South Carolina. This material is supported in part by NIH R01 GM43825 (to YAH) and a VA Merit Award, Office of Research and Development, Department of Veterans Affairs, NIH R01 AG016583 (to LMO). L. A. C. would like to acknowledge support from the Department of Veteran's Affairs Merit Review Entry Program. A part of this study was financially supported by the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on the screening and counseling by the Atomic Energy Commission.
Abbreviations
- IPC
inositol phosphorylceramide
- MIPC
mannosylinositol phosphorylceramide
- M(IP)2C
mannosyldiinositol phosphorylceramide
- YPD
yeast/peptone/dextrose
- YPGE
yeast/peptone/glycerol/ethanol
- SEM
standard error of mean
- ER
endoplasmic reticulum
- IS
internal standard
- a-HO-
α-hydroxylated
- Cer
ceramide
- DHSph
dihydrosphingosine
- Phyto-Sph
phytosphingosine
- dhCn-Cer
dihydroceramide with Cn fatty acid
- phytoCn-Cer
phytoceramide with Cn fatty acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hannun YA, Obeid LM. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 2.Smith WL, Merrill AH., Jr. Sphingolipid metabolism and signaling minireview series. J. Biol. Chem. 2002;277:25841–25842. doi: 10.1074/jbc.R200011200. [DOI] [PubMed] [Google Scholar]
- 3.Dickson RC, Lester RL. Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1999;1438:305–321. doi: 10.1016/s1388-1981(99)00068-2. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell. Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds CP, Maurer BJ, Kolesnick RN. Ceramide synthesis and metabolism as a target for cancer therapy. Cancer Lett. 2004;206:169–180. doi: 10.1016/j.canlet.2003.08.034. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins GM, Hannun YA. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:8574–8581. doi: 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- 7.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15:312–318. doi: 10.1016/j.tcb.2005.04.006. 2005. [DOI] [PubMed] [Google Scholar]
- 9.Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur. J. Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- 10.Patton JL, Lester RL. The phosphoinositol sphingolipids of Saccharomyces cerevisiae are highly localized in the plasma membrane. J. Bacteriol. 1991;173:3101–3108. doi: 10.1128/jb.173.10.3101-3108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futerman AH. Intracellular trafficking of sphingolipids: relationship to biosynthesis Biochim. Biophys. Acta. 2006;1758:1885–18892. doi: 10.1016/j.bbamem.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Bionda C, Portoukalian J, Schmitt D, Rodriguez-Lafrasse C, Ardail D. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 2004;382:527–533. doi: 10.1042/BJ20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birbes H, El Bawab S, Hannun YA, Obeid LM. Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. FASEB J. 2001;15:2669–2679. doi: 10.1096/fj.01-0539com. [DOI] [PubMed] [Google Scholar]
- 14.Kashkar H, Wiegmann K, Yazdanpanah B, Haubert D, Kronke M. M. Acid sphingomyelinase is indispensable for UV light-induced Bax conformational change at the mitochondrial membrane. J. Biol. Chem. 2005;280:20804–20813. doi: 10.1074/jbc.M410869200. [DOI] [PubMed] [Google Scholar]
- 15.Novgorodov SA, Szulc ZM, Luberto C, Jones JA, Bielawski J, Bielawska A, Hannun YA, Obeid LM. Positively charged ceramide is a potent inducer of mitochondrial permeabilization. J. Biol. Chem. 2005;280:16096–16105. doi: 10.1074/jbc.M411707200. [DOI] [PubMed] [Google Scholar]
- 16.Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J. Biol. Chem. 1997;272:24154–24158. doi: 10.1074/jbc.272.39.24154. [DOI] [PubMed] [Google Scholar]
- 17.Siskind LJ, Kolesnick RN, Colombini M. Ceramide channels increase the permeability of the mitochondrial outer membrane to small proteins. J. Biol. Chem. 2002;277:26796–26803. doi: 10.1074/jbc.M200754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimeno H, Soeda S, Sakamoto M, Kouchi T, Kowakame T, Kihara T. Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine liver mitochondrion-rich fraction. Lipids. 1998;33:601–605. doi: 10.1007/s11745-998-0246-2. [DOI] [PubMed] [Google Scholar]
- 19.El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters JJ, Hannun YA. Molecular cloning and characterization of a human mitochondrial ceramidase. J. Biol. Chem. 2000;275:21508–21513. doi: 10.1074/jbc.M002522200. [DOI] [PubMed] [Google Scholar]
- 20.Ardail D, Popa I, Bodennec J, Louisot P, Schmitt D, Portoukalian J. The mitochondria-associated endoplasmic-reticulum subcompartment (MAM fraction) of rat liver contains highly active sphingolipid-specific glycosyltransferases. Biochem. J. 2003;371:1013–1019. doi: 10.1042/BJ20021834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi K, Hata K, Koseki K, Shiozaki K, Akita H, Wada T, Moriya S, Miyagi T. Evidence for mitochondrial localization of a novel human sialidase (NEU4). Biochem. J. 2005;390:85–93. doi: 10.1042/BJ20050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Toman RE, Goparaju SK, Maceyka M, Nava VE, Sankala H, Payne SG, Bektas M, Ishii I, Chun J, Milstien S, Spiegel S. Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. J. Biol. Chem. 2003;278:40330–40336. doi: 10.1074/jbc.M304455200. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA. Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature. 2005;433:425–430. doi: 10.1038/nature03232. [DOI] [PubMed] [Google Scholar]
- 24.Vaena de Avalos S, Okamoto Y, Hannun YA. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:11537–11545. doi: 10.1074/jbc.M309586200. [DOI] [PubMed] [Google Scholar]
- 25.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:39793–39798. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 26.Okamoto Y, Vaena De Avalos S, Hannun YA. Structural requirements for selective binding of ISC1 to anionic phospholipids. J. Biol. Chem. 2002;277:46470–46677. doi: 10.1074/jbc.M207779200. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto Y, Vaena de Avalos S, Hannun YA. Functional analysis of ISC1 by site-directed mutagenesis. Biochemistry. 2003;42:7855–7862. doi: 10.1021/bi0341354. [DOI] [PubMed] [Google Scholar]
- 28.Cowart LA, Okamoto Y, Lu X, Hannun YA. Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem. J. 2005;393:733–740. doi: 10.1042/BJ20050643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:7170–7177. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- 30.Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- 31.Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisinger C, Sommer T, Pfanner N. Purification of Saccharomcyes cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 2000;287:339–342. doi: 10.1006/abio.2000.4868. [DOI] [PubMed] [Google Scholar]
- 33.Ohlmeier S, Kastaniotis AJ, Hiltunen JK, Bergmann U. The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J. Biol. Chem. 2004;279:3956–3979. doi: 10.1074/jbc.M310160200. [DOI] [PubMed] [Google Scholar]
- 34.Bielawski J, Szulc ZM, Hannun YA, Bielawska A. Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods. 2006;39:82–91. doi: 10.1016/j.ymeth.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Ryan MT, Voos W, Pfanner N. Assaying protein import into mitochondria. Methods in Cell Biology. 2001;65:202–204. doi: 10.1016/s0091-679x(01)65012-x. [DOI] [PubMed] [Google Scholar]
- 36.Ostrander DB, Zhang M, Mileykovskaya E, Rho M, Dowhan W. W. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J. Biol. Chem. 2001;276:25262–25272. doi: 10.1074/jbc.M103689200. [DOI] [PubMed] [Google Scholar]
- 37.Daum G. Lipids of mitochondria. Biochim. Biophys. Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 38.Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 39.Zhong Q, Gohil VM, Ma L, Greenberg ML. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 2004;279:32294–32300. doi: 10.1074/jbc.M403275200. [DOI] [PubMed] [Google Scholar]
- 40.Demasi AP, Pereira GA, Netto LE. Cytosolic thioredoxin peroxidase I is essential for the antioxidant defense of yeast with dysfunctional mitochondria. FEBS Lett. 2001;509:430–434. doi: 10.1016/s0014-5793(01)03215-x. [DOI] [PubMed] [Google Scholar]
- 41.Slonimski PP, Perrodin G, Croft JH. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites”. Biochem. Biophys. Res. Commun. 1968;30:232–239. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- 42.Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165:35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickson RC, Sumanasekera C, Lester CRL. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid. Res. 2006;45:447–465. doi: 10.1016/j.plipres.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Uemura S, Kihara A, Iwaki S, Inokuchi J, Igarashi Y. Regulation of the transport and protein levels of the inositol phosphorylceramide mannosyltransferases Csg1 and Csh1 by the Ca2+-binding protein Csg2. J Biol Chem. 2007;282:8613–8621. doi: 10.1074/jbc.M606649200. [DOI] [PubMed] [Google Scholar]
- 46.Uemura S, Kihara A, Inokuchi J, Igarashi Y. Csg1p and newly identified Csh1p function in mannosylinositol phosphorylceramide synthesis by interacting with Csg2p. J Biol Chem. 2003;278:45049–45055. doi: 10.1074/jbc.M305498200. [DOI] [PubMed] [Google Scholar]
- 47.Levine TP, Wiggins CA, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardail D, Popa I, Alcantara K, Pons A, Zanetta JP, Louisot P, Thomas L, Portoukalian J. Occurrence of ceramides and neutral glycolipids with unusual long-chain base composition in purified rat liver mitochondria. FEBS Lett. 2001;488:160–164. doi: 10.1016/s0014-5793(00)02332-2. [DOI] [PubMed] [Google Scholar]
- 49.Sperka-Gottlieb CD, Hermetter A, Paltauf F, Daum G. Lipid topology and physical properties of the outer mitochondrial membrane of the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1988;946:227–234. doi: 10.1016/0005-2736(88)90397-5. [DOI] [PubMed] [Google Scholar]
- 50.Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simbeni R, Pon L, Zinser E, Paltauf F, Daum G. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J. Biol. Chem. 1991;266:10047–10049. [PubMed] [Google Scholar]
- 52.Ardail D, Lerme F, Louisot P. Involvement of contact sites in phosphatidylserine import into liver mitochondria. J. Biol. Chem. 1991;266:7978–7981. [PubMed] [Google Scholar]
- 53.Simbeni R, Paltauf F, Daum G. Intramitochondrial transfer of phospholipids in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:281–285. [PubMed] [Google Scholar]
- 54.Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hovius R, Lambrechts H, Nicolay K, de Kruijff B. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim. Biophys. Acta. 1990;1021:217–226. doi: 10.1016/0005-2736(90)90036-n. [DOI] [PubMed] [Google Scholar]
- 56.Hirschberg K, Rodger J, Futerman AH. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 1993;290:751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao C, Xu R, Bielawska A, Obeid LM. Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem. 2000;275:6876–6884. doi: 10.1074/jbc.275.10.6876. [DOI] [PubMed] [Google Scholar]
- 58.Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. Cloning and characterization of a Saccharomyces cerevisiae alkaline ceramidase with specificity for dihydroceramide. J. Biol. Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- 59.Bae TJ, Kim MS, Kim JW, Kim BW, Choo HJ, Lee JW, Kim KB, Lee CS, Kim JH, Chang SY, Kang CY, Lee SW, Ko YG. Lipid raft proteome reveals ATP synthase complex in the cell surface. Proteomics. 2004;4:3536–3548. doi: 10.1002/pmic.200400952. [DOI] [PubMed] [Google Scholar]
- 60.Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. U S A. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghafourifar P, Klein SD, Schucht O, Schenk U, Pruschy M, Rocha S, Richter C. Ceramide induces cytochrome c release from isolated mitochondria. Importance of mitochondrial redox state. J. Biol. Chem. 1999;274:6080–6084. doi: 10.1074/jbc.274.10.6080. [DOI] [PubMed] [Google Scholar]
- 63.Kashkar H, Wiegmann K, Yazdanpanah B, Haubert D, Kronke M. Acid sphingomyelinase is indispensable for UV light-induced Bax conformational change at the mitochondrial membrane. J. Biol. Chem. 2005;280:20804–20813. doi: 10.1074/jbc.M410869200. [DOI] [PubMed] [Google Scholar]