Abstract
The 9ORF1 gene encodes an adenovirus E4 region oncoprotein that requires a C-terminal region for transforming activity. Screening a λgt11 cDNA expression library with a 9ORF1 protein probe yielded a novel cellular PDZ domain-containing protein, 9BP-1, which binds to wild-type, but not a transformation-defective, C-terminal, mutant 9ORF1 protein. The fact that PDZ domains complex with specific sequences at the free C-terminal end of some proteins led to the recognition that the 9ORF1 C-terminal region contained such a consensus-binding motif. This discovery prompted investigations into whether the 9ORF1 protein associates with additional cellular proteins having PDZ domains. It was found that the 9ORF1 protein interacts directly, in vitro and in vivo, with the PDZ domain-containing protein hDlg/SAP97 (DLG), which is a mammalian homolog of the Drosophila discs large tumor suppressor protein and which also binds the adenomatous polyposis coli tumor suppressor protein. Of interest, in forming complexes, the 9ORF1 protein preferentially associated with the second PDZ domain of DLG, similar to adenomatous polyposis coli protein. Human T cell leukemia virus type 1 Tax and most oncogenic human papillomavirus E6 oncoproteins also possessed PDZ domain-binding motifs at their C termini and, significantly, human T cell leukemia virus type 1 Tax and human papillomavirus 18 E6 proteins bound DLG in vitro. Considering the requirement of the 9ORF1 C-terminal region in transformation, these findings suggest that interactions with the cellular factor DLG may contribute to the tumorigenic potentials of several different human virus oncoproteins.
Human adenoviruses are organized into six subgroups, A through F, and in people cause primarily respiratory, gastrointestinal, and eye infections (1). After infection of rodents, however, subgroup A and B and some subgroup D adenoviruses are tumorigenic. In animals, subgroup A and B adenoviruses produce undifferentiated sarcomas at the site of virus injection, and the viral E1 region, consisting of the E1A and E1B genes, is both necessary and sufficient for this tumorigenicity. The transforming potentials of the nuclear E1A and E1B oncoproteins derive, at least in part, from an ability to complex with and inactivate the cellular tumor suppressor proteins pRB and p53, respectively (2).
Compared with the subgroup A and B adenoviruses, subgroup D adenovirus type 9 (Ad9) is unique in eliciting only estrogen-dependent mammary tumors in female rats (3) and requiring the viral E4 region ORF1 gene (9ORF1) for oncogenicity (4, 5). The 9ORF1 gene codes for a 14-kDa cytoplasmic transforming protein (5, 6), and three separate regions of this viral polypeptide are important for transforming potential in cells (7), including a C-terminal region that mediates direct binding to several unidentified cellular proteins (p220, p180, p160, p155, p140/p130; R.S.W. and R.T.J., unpublished work). To reveal the molecular mechanisms of the novel 9ORF1 oncoprotein, we sought to identify these 9ORF1-associated cellular polypeptides.
We report here that the 9ORF1 protein possesses a C-terminal PDZ domain-binding motif that is necessary for binding to the PDZ domains of the cellular factor hDLg/SAP97 (DLG) (8, 9). This finding is significant because DLG is a mammalian homolog of the Drosophila discs large tumor suppressor protein (10) and, in mammalian cells, complexes with the product of the adenomatous polyposis coli (APC) tumor suppressor gene (11), which is mutated in most familial and sporadic colon cancers (12). Additionally, we show that the human T cell leukemia virus type 1 (HTLV-1) Tax and human papillomavirus (HPV) 18 E6 oncoproteins also encode consensus C-terminal PDZ domain-binding motifs and similarly bind to DLG. Therefore, DLG may represent an important cellular target for several different human virus oncoproteins.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents.
Portions of the SAP97 (rat DLG) ORF were PCR amplified with pfu DNA polymerase (Stratagene) from a pRK174-SAP97 plasmid (provided by C. C. Garner) (8) template using the following primer sets flanked by BamHI and EcoRI sites: NT, (a) CTC GGA TCC ATG CCG GTC CGG AAG CAA and (b) CTC GAA TTC CTC ATA TTC ATA ATC TGC; 3PDZ, (c) CTC GGA TCC GCA GAT TAT GAA TAT GAG and (d) CTC GAA TTC GGT TCG GAG AGA CCC TGA; SH3/GuK, (e) CTC GAA TTC TTT CAG GGT CTC TCC GAA CC and (f) CTC GAA TTC TCA TAA TTT TTC TTT TGC TGG GAC CC; PDZ1–2, (c) and (g) CTC GAA TTC ACT TGT TGG TTT TGC CGC; PDZ2–3, (d) and (h) CTC GGA TCC CCT TCA GAA AAA ATC ATG; PDZ1, (c) and (i) CTC GAA TTC GAA GGC CTT CCG CCT TTT; PDZ2, (g) and (h); and PDZ3, (d) and (j) CTC GGA TCC ATG TAT ATA AAT GAT GGC. Full length and PCR-generated SAP97 fragments, as well as wild type and mutant E4 ORF1 genes, were cloned in-frame with the glutathione S-transferase (GST) gene of pGEX-2T or pGEX-2TK at the BamHI and EcoRI sites. PCR products were verified to be correct by partial or complete sequence analysis. pGEX-2T plasmids containing HPV-11 and HPV-18 E6, HTLV-1 Tax, and HIV-1 Tat genes were gifts from P. Howley, S. Marriott, and A. Rice, respectively. GST fusion proteins were expressed in Escherichia coli and purified on glutathione–Sepharose beads by standard methods (13). The λgt11 murine pancreatic cell cDNA library was provided by S. Tsai.
Cells and Cell Extracts.
The rat embryo fibroblast cell line CREF, human osteosarcoma-derived cell line TE85, and G418-selected CREF cell pools containing an empty expression plasmid (vector) or expression plasmid encoding the wild-type or mutIII-A 9ORF1 gene were maintained as described (7, 14). For the preparation of cell lysates, cells were harvested, washed with PBS, and incubated in ≈5 vol of RIPA buffer [50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1% (vol/vol) Nonidet P-40/0.5% (wt/vol) sodium desoxycholate/0.1% (wt/vol) SDS] containing protease inhibitors (300 μg/ml phenylmethylsulfonyl fluoride and 6 μg/ml each of aprotinin and leupeptin) for 10 min on ice. Cell lysates were cleared of cell debris by centrifugation at 10,000 × g for 10 min. Protein concentrations were determined by the Bradford method.
Protein Blotting Assays.
Protein blotting assays were performed with lambda phage plaques immobilized on nitrocellulose membranes or proteins separated by SDS/PAGE and transferred to poly(vinylidene fluoride) membranes (15). Membranes were blocked in TBST buffer [50 mM Tris⋅HCl, pH 7.5/200 mM NaCl/0.2% (vol/vol) Tween 20] containing 5% nonfat dried milk, incubated in blotting buffer (0.1% nonfat dry milk and 100 μg/ml unlabeled GST protein in TBST) containing [32P]-labeled GST-9ORF1 protein probe (5 × 105 cpm/ml) for 12 h at 4°C, washed extensively with RIPA buffer, and developed by autoradiography. GST fusion protein probes were generated by incubating ≈3 μg of affinity-purified GST fusion protein bound to beads in reaction buffer [20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/12 mM MgCl2/20 μCi [32P-γ-ATP] (6000 Ci/mmol)] containing 10 units of protein kinase A (Sigma) for 30 min on ice. Beads were washed extensively with RIPA buffer and eluted in elution buffer [20 mM Tris⋅HCl, pH 8.0/100 mM NaCl/1 mM EDTA/0.5% (vol/vol) Nonidet P-40/40 mM glutathione].
GST Pull-Down Reactions, Immunoprecipitations, and Immunoblots.
GST pull-down reactions were performed by incubating cell lysates with ≈5 μg of affinity-purified GST fusion protein attached to glutathione–Sepharose beads. Immunoprecipitations were carried out by incubating cell lysates with 9ORF1 antiserum or affinity-purified DLG antibodies (supplied by M. Sheng) for 3 h and with 30 μl of protein A–Sepharose beads for 1 h on ice. After extensive RIPA buffer washes, cellular proteins bound to the respective beads were separated by 7.5% SDS/PAGE and transferred to a poly(vinylidene fluoride) membrane. Membranes were immunoblotted with either 9ORF1 rabbit polyclonal antiserum (5) or DLG antibodies (supplied by D. Branton and C. C. Garner) as described (6).
Database Searches.
Protein database searches for polypeptides encoding C-terminal PDZ domain-binding motifs were performed using the command (S, T)X(V, I)> with the FindPatterns algorithm of the Genetics Computer Group (Madison, WI) software package.
RESULTS AND DISCUSSION
The 9ORF1 Protein Possesses a Consensus C-Terminal PDZ Domain-Binding Motif.
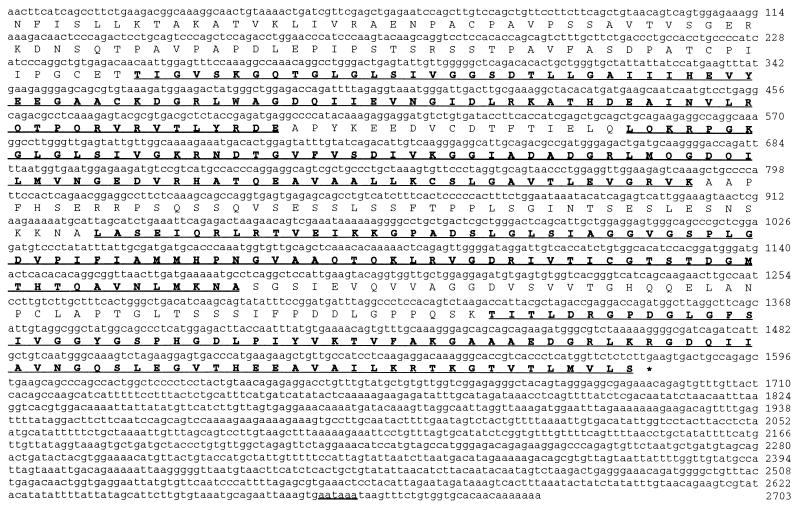
To identify cellular factors interacting with the 9ORF1 C-terminal region, we screened a λgt11 cDNA expression library, derived from a murine pancreatic cell line that expresses 9ORF1-associated cellular proteins, with a radiolabeled GST-9ORF1 fusion protein probe. A single phage plaque was isolated that reacted with the wild-type but not with a mutIII-A 9ORF1 probe (data not shown). As a result of a C-terminal mutation, the mutIII-A 9ORF1 protein fails to transform cells (7) or bind any of the 9ORF1-associated cellular proteins (Table 1; unpublished results). The isolated phage contained a 2.7-kb partial cDNA coding for the C-terminal 526 amino acid residues of a novel cellular protein, designated 9BP-1 (9ORF1-Binding Protein 1) (Fig. 1). Although not found in the sequence databases, the recovered portion of 9BP-1 shared sequence similarity with many PDZ domain-containing proteins and, upon inspection of homologous regions, was found to consist of four PDZ domains separated by segments of unique protein sequence (Fig. 1).
Table 1.
Adenovirus E4 ORF1 transforming proteins possess consensus PDZ domain-binding motifs at their C termini*
| PDZ domain-binding motif | Position
|
|||||
|---|---|---|---|---|---|---|
| −3 −2 −1 0
| ||||||
| Transforming activity† | X | (S/T) | X | (V/I) | ||
| 9ORF1 | +++++ | A | T | L | V | |
| mutIII-A | − | A | P | |||
| mutIII-B | − | A | T | L | V | SACFL |
| mutIII-C | + | D | T | L | V | |
| mutIII-D | + | A | T | P | V | |
| T123A | ND | A | A | L | V | |
| T123D | ND | A | D | L | V | |
| V125A | ND | A | T | L | A | |
| 12ORF1 | +++++ | A | S | L | I | |
| 3ORF1 | +++++ | A | T | M | I | |
| 5ORF1 | +++++ | A | S | N | V | |
| APC | N/A | V | T | S | V | |
The four C-terminal amino acid residues of each wild-type protein are shown. Alterations of mutant 9ORF1 proteins are depicted as outlined amino acid residues.
Figure 1.
Nucleotide and predicted amino acid sequences of the 2.7-kb partial murine 9BP-1 cDNA recovered from the 9BP-1 phage. The underlined and bold 82–84 amino acid residue segments designate four protein regions having sequence similarity to PDZ domains. Underlined nucleotides denote the putative 9BP-1 polyadenylylation signal.
PDZ domain-containing proteins normally localize to specialized sites of cell–cell contact, such as tight junctions in epithelial cells and synaptic densities in neurons, and are thought to function in signal transduction (16, 17). Like SRC homology region 2 (SH2), SH3, and phosphotyrosine-binding domains, PDZ domains are modular protein units that mediate protein–protein interactions, and certain PDZ domains bind directly to the consensus sequence S/T-X-V/I (X denotes any amino acid) at the free C-terminal end of some proteins (18–20). Significantly, 9ORF1 and other adenovirus E4 ORF1 transforming proteins (12ORF1, 3ORF1, and 5ORF1) (14) possessed this consensus, C-terminal, PDZ domain-binding motif (Table 1). Moreover, mutations within or adjacent to this motif decrease the ability of 9ORF1 to transform cells (mutIII-A, mutIII-B, mutIII-C, and mutIII-D; Table 1) (7) and bind some or all 9ORF1-associated cellular proteins. These observations led us to predict that, like 9BP-1, other cellular factors found to complex with the 9ORF1 protein through its C-terminal region would contain PDZ domains.
The 9ORF1 Protein Complexes with the PDZ Domain-Containing Cellular Protein DLG in Vitro.
A search for known PDZ domain-containing proteins having an established or suspected role in neoplasia revealed one particularly attractive candidate protein, hDlg/SAP97 (DLG). DLG is the mammalian homolog for the Drosophila discs large tumor suppressor protein Dlg-A. Both DLG and Dlg-A are members of the membrane-associated guanylate kinase (MAGUK) family of proteins that contain, in addition to PDZ domains, an SH3 domain and a region with homology to guanylate kinases (16). In Drosophila imaginal disc epithelia, Dlg-A localizes to septate junctions, the equivalent of tight junctions in mammalian cells, and homozygous Dlg-A mutations lead to disruption of cell junctions, shape, and polarity, as well as neoplastic growth (21). For the mammalian homolog DLG, expression is found in many tissues in vivo, as well as cell lines derived from fibroblasts and epithelial, B, and T cells, and, like Dlg-A, DLG localizes to regions of cell–cell contact in epithelial cells (8, 9). As another possible link to neoplasia, DLG complexes with the tumor suppressor protein APC in mammalian cells (11). Of interest, this interaction requires a PDZ domain of DLG and the APC C terminus, which encodes a consensus PDZ domain-binding motif (Table 1). Moreover, DLG migrates at 140-kDa in protein gels (8, 11), similar to the unidentified p140/p130 9ORF1-associated cellular proteins. These facts compelled us to investigate whether the 9ORF1 oncoprotein binds to this cellular factor.
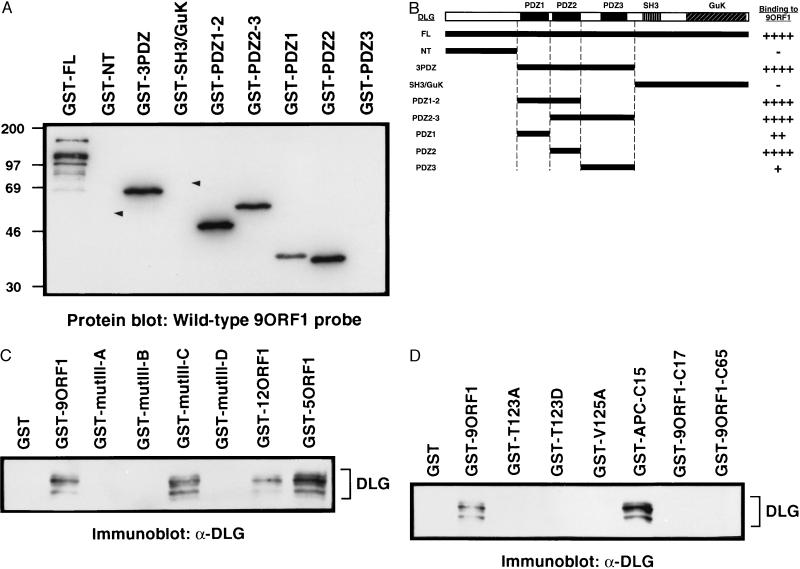
Because, like 9BP-1, 9ORF1-associated cellular proteins immobilized on a membrane bind specifically to a wild-type GST-9ORF1 protein probe (R.S.W. and R.T.J., unpublished results), this approach was chosen initially to examine a possible interaction between the 9ORF1 and DLG polypeptides. Significantly, in such experiments, electrophoretically separated, membrane-immobilized, full length DLG fusion protein, but not GST alone (data not shown), reacted with the radiolabeled wild-type 9ORF1 probe (Fig. 2A). In contrast, full length DLG fusion protein did not react with a mutIII-A 9ORF1 probe (data not shown), indicating that the interaction between the wild-type 9ORF1 and DLG proteins was specific and mediated by the 9ORF1 C-terminal domain. To identify which region of DLG was binding to the wild-type 9ORF1 protein, we separated DLG into three protein fragments and analyzed each fragment fused to GST in similar assays. We found that the DLG fragment containing all three PDZ domains (GST-3PDZ) interacted with the 9ORF1 probe, but the unique N-terminal (GST-NT) and SH3 and guanylate kinase domain-containing C-terminal (GST-SH3/GuK) fragments did not (Fig. 2A). Further analyses to delineate which DLG PDZ domain interacted with the 9ORF1 protein revealed that the 9ORF1 probe bound most strongly to fragments containing the second DLG PDZ domain (GST-PDZ1–2, GST-PDZ2–3, GST-PDZ2) yet also reacted with the first (GST-PDZ1) and weakly with the third (GST-PDZ3) DLG PDZ domains individually (Fig. 3A). Of interest, APC also binds to the second PDZ domain of DLG (11). These results, summarized in Fig. 2B, indicated that the wild-type 9ORF1 protein binds directly to DLG and, as predicted, this interaction is mediated by the PDZ domains of this cellular protein.
Figure 2.
Association of the 9ORF1 oncoprotein with the cellular factor DLG in vitro. (A) Binding of a wild-type GST-9ORF1 protein probe to bacterially expressed membrane-immobilized GST-DLG proteins (1 μg) in a protein blotting assay. Arrowheads show locations of GST-NT and GST-SH3/GuK proteins that did not bind to the GST-9ORF1 probe. The multiple bands seen with full length DLG fusion protein are probably proteolytic products of this large fusion protein. (B) An illustration of the DLG protein fragments used in A and summary of their corresponding 9ORF1 protein-binding activities. ++++, strong binding activity; ++, moderate binding activity; +, weak binding activity; −, no detectable binding activity. (C) Interaction of wild-type and transformation-defective C-terminal mutant 9ORF1 proteins with cellular DLG in vitro. GST pull-down reactions with the indicated GST fusion proteins were performed with 1 mg of protein from CREF cell lysates, and recovered cellular proteins were immunoblotted with rabbit polyclonal antiserum to DLG (9). Protein bands were visualized with an enhanced chemiluminescence detection system. (D) Interaction of C-terminal mutant 9ORF1 proteins and C-terminal 9ORF1 fragments with cellular DLG in vitro. GST pull-down reactions were performed as described in C. See Table 1 for description of mutant 9ORF1 proteins.
Figure 3.
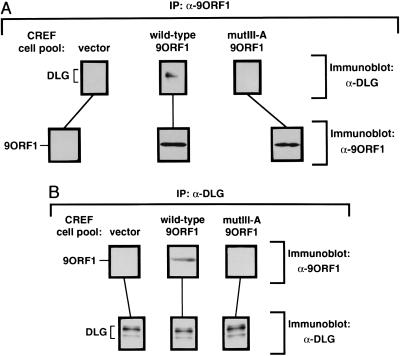
Association of the 9ORF1 oncoprotein with DLG in vivo. (A) Coimmunoprecipitation of cellular DLG with the 9ORF1 protein. Two milligrams of protein from cell lysates prepared from the indicated CREF cell pool were subjected to immunoprecipitation with 2.5 μl of 9ORF1 rabbit polyclonal antiserum. Precipitated proteins were separated by SDS/PAGE, transferred to a poly(vinylidene fluoride) membrane, and immunoblotted with either DLG rabbit polyclonal antiserum (9) (upper three panels) or 9ORF1 antiserum (lower three panels). (B) Coimmunoprecipitation of the 9ORF1 protein with cellular DLG. These experiments were performed as described in A except that cellular lysates were immunoprecipitated with 0.4 μg of affinity-purified DLG rabbit polyclonal antibodies (34). Membranes were immunoblotted with either 9ORF1 antiserum (upper three panels) or DLG antiserum (lower three panels). IP, immunoprecipitation.
We also demonstrated that the GST-9ORF1 protein interacts specifically with authentic cellular DLG. This was accomplished by incubating wild-type GST-9ORF1 protein with whole cell lysates and immunoblotting the recovered cellular proteins with a polyclonal antiserum against DLG. The wild-type GST-9ORF1 protein, as well as the related wild-type GST-12ORF1 and GST-5ORF1 proteins (14), bound to DLG in the cellular extracts whereas GST alone did not (Fig. 2C). The detection of several ≈140-kDa DLG species in these experiments was presumably due to the fact that it exists in four different alternatively spliced isoforms (9). These results were confirmed independently with an mAb to DLG (8) (data not shown). In the same experiments, we also included four transformation-defective C-terminal mutant 9ORF1 proteins (mutIII-A, mutIII-B, mutIII-C, mutIII-D; see Table 1) (7). With the exception of mutIII-C, all of these mutant 9ORF1 proteins failed to complex with DLG (Fig. 2C). The mutIII-A and mutIII-B proteins lack detectable transforming activity and binding to any 9ORF1-associated cellular proteins whereas the mutIII-C and mutIII-D proteins retain weak transforming activity and binding to some 9ORF1-associated cellular proteins (ref. 7 and R.S.W. and R.T.J., unpublished results). The mutIII-C protein uniquely preserves the capacity to complex with p140/p130 (DLG) but not other 9ORF1-associated cellular proteins, and, conversely, the mutIII-D protein fails to interact with p140/p130 (DLG) but binds other 9ORF1-associated cellular proteins. Taken together, these results suggested that the 9ORF1 oncoprotein must complex with DLG, as well as one or more other 9ORF1-associated cellular proteins, to sustain full transforming potential.
Similar protein binding assays were carried out to further characterize the 9ORF1 C-terminal domain. For related PDZ domain-binding motifs, mutation of highly conserved amino acid residues at position −2 or position 0 from the C terminus results in loss of protein binding activity (20). Therefore, we introduced point mutations at equivalent amino acid residue positions of the 9ORF1 protein (T123A, T123D, V125A; see Table 1) to determine whether DLG-binding activity would likewise be eliminated. Concordant with results for other PDZ domain-binding motifs, such alterations significantly diminished binding of these mutant 9ORF1 polypeptides to DLG (Fig. 2D). In addition, because the 10–15 C-terminal amino acid residues of other PDZ domain-binding proteins retain protein binding activity (11, 19), we investigated also whether 9ORF1 C-terminal protein fragments would preserve reactivity toward DLG. Whereas the C-terminal 15 amino acid residues of APC (GST-APC-C15) possessed potent DLG-binding activity, the C-terminal 17 and 65 amino acid residues of the 9ORF1 protein (GST-9ORF1-C17 and GST-9ORF1-C65) failed to demonstrate detectable interactions with DLG (Fig. 2D). These C-terminal 9ORF1 fragments also either failed to bind or showed reduced binding to some of the other 9ORF1-associated cellular proteins (R.S.W. and R.T.J., unpublished results). Thus, while sharing many characteristics with related PDZ domain-binding motifs, the 9ORF1 C-terminal region exhibited the novel property of requiring an intact 9ORF1 polypeptide for retention of wild-type protein binding activity.
The 9ORF1 Protein Complexes with the PDZ Domain-Containing Cellular Protein DLG in Vivo.
Although the results described thus far established that the 9ORF1 protein binds to DLG in vitro, it was essential to show that these polypeptides also form complexes in vivo. For this purpose, we subjected various cellular lysates to immunoprecipitation analyses with 9ORF1 antiserum and immunoblotted the recovered protein precipitates with anti-DLG polyclonal serum. In these experiments, the 9ORF1 antibodies coprecipitated DLG from wild-type 9ORF1-expressing cells but not from normal cells or from cells expressing mutIII-A 9ORF1 (Fig. 3A). A reciprocal analysis, in which cellular lysates were immunoprecipitated with an affinity-purified DLG polyclonal serum and recovered protein precipitates were immunoblotted with 9ORF1 antiserum, confirmed these findings because the DLG antibodies coprecipitated 9ORF1 protein from wild-type 9ORF1-expressing cells but not from normal cells or from cells expressing mutIII-A 9ORF1 (Fig. 3B). In addition, DLG also coimmunoprecipitated with 9ORF1 protein expressed in an Ad9-induced rat mammary tumor cell line (data not shown). These results indicated that the wild type, but not a C-terminal mutant 9ORF1 protein, forms complexes with DLG in vivo.
The HTLV-1 Tax and HPV-18 E6 Oncoproteins Possess Consensus C-Terminal PDZ Domain-Binding Motifs and Bind DLG in Vitro.
Seemingly unrelated viral oncoproteins may share common strategies for cellular transformation, a concept illustrated by the fact that the simian virus 40 large T antigen, the papillomavirus E6 and E7 proteins, and the adenovirus E1A and E1B proteins similarly target the cellular tumor suppressor proteins pRB and p53 (22). Consequently, we considered the possibility that other viral oncoproteins, in addition to the 9ORF1 protein, might also bind DLG. This idea was initially explored by searching sequence databases for polypeptides encoding a consensus PDZ domain-binding motif at their free C-terminal ends. From this analysis, we discovered that the Tax oncoprotein from human T cell leukemia virus type 1 (HTLV-1) and many different E6 oncoproteins from HPVs of the genital tract also possessed a consensus C-terminal PDZ domain-binding motif (Table 2). The retrovirus HTLV-1 is the etiological agent of the aggressive and often fatal adult T cell leukemia (23) whereas certain HPVs are associated with the majority of cervical cancers (24). Of interest, with the exception of those encoded by HPV-16 and HPV-33, all E6 proteins from HPVs generating genital lesions with a risk for malignant progression (HPV types 16, 18, 30, 31, 33, 35, 39, 45, 51, 52, 56, 58) contained the putative PDZ domain-binding motif (25).
Table 2.
HTLV-1 Tax and many HPV E6 oncoproteins have consensus PDZ domain-binding motifs at their C termini*
| PDZ domainbinding motif | Position
|
|||
|---|---|---|---|---|
| −3 −2 −1 0
| ||||
| X | (S/T) | X | (V/I) | |
| HTLV-1 Tax | E | T | E | V |
| HIV-1 Tat | G | P | K | E |
| HPV E6 types | ||||
| 11 | D | L | L | P |
| 18, 26, 31, 39, 45, 51 | E | T | Q | V |
| 30 | E | T | A | V |
| 34 | A | T | V | V |
| 35 | E | T | E | V |
| 52 | V | T | Q | V |
| 53 | E | S | A | V |
| 56 | E | S | T | V |
| 58 | Q | T | Q | V |
The C-terminal four amino acid residues of each viral protein are shown. Note that HIV-1 Tat and, with the exception of those encoded by HPV-26, HPV-34, and HPV-53, E6 proteins from low risk HPVs (e.g., HPV-11 E6) do not encode the indicated consensus C-terminal PDZ domain-binding motif.
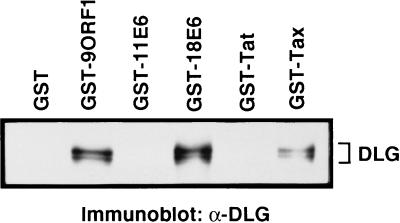
These observations prompted experiments to address whether HTLV-1 Tax and HPV E6 oncoproteins also would bind DLG. For this purpose, we examined Tax and one representative E6 protein, from HPV-18, which is designated as a high risk virus because of its strong association with cervical cancer. The HPV-18 E6 protein (18E6) also has the identical C-terminal sequence as five other E6 proteins, including those from two additional high risk viruses, HPV-31 and HPV-45 (Table 2). To assay binding to cellular DLG, we incubated GST-Tax and GST-18E6 proteins with protein extracts from a human cell line and immunoblotted the recovered proteins with anti-DLG polyclonal serum. Significantly, similar to GST-9ORF1, both GST-Tax and GST-18E6 specifically associated with cellular DLG (Fig. 4). GST alone as well as HIV-1, Tat (GST-Tat), and low risk HPV-11 E6 (GST-11E6) fusion proteins, which lacked the consensus C-terminal PDZ domain-binding motif (Table 2), failed to demonstrate detectable DLG-binding activity in these experiments (Fig. 4). Because the HTLV-1 Tax and HPV E6 proteins, aside from having nuclear components, also localize to the cytoplasm of cells similar to the 9ORF1 protein (26, 27), in vivo interactions with DLG seem possible. Moreover, DLG is expressed in both human T and cervical epithelial cell lines (9) (unpublished results), cell types infected by HTLV-1, and genital HPVs, respectively. Although transformation by the Tax or E6 oncoproteins is known to require binding to the cellular factors CREB, CBP, and NFKB2 (28–31) or p53, E6-AP, and E6BP (25, 26), respectively, the results suggest that these viral oncoproteins also complex with DLG. Thus, considering the findings with the 9ORF1 protein, DLG binding may also contribute to the transforming potential of these viral oncoproteins. In support of this idea, the Tax C-terminal 23 amino acid residues, which presumably mediate binding to DLG, have been implicated in Tax-mediated transformation (32).
Figure 4.
Association of HTLV-1 Tax and HPV E6 oncoproteins with DLG in vitro. GST pull-down reactions with the indicated GST fusion protein were performed as described in the legend to Fig. 2 except that 500 μg of protein from human TE85 cell lysate was used per reaction.
PDZ domains serve to recruit and network plasma membrane and cytoskeletal proteins to regions of cell–cell contact (16, 17). Moreover, PDZ domain-containing polypeptides, like DLG, normally possess additional protein motifs, including SH3, guanylate kinase-like, pleckstrin, protein tyrosine phosphatase, or Ca+/calmodulin protein kinase II domains (33), suggesting that these cellular factors function as components of novel signal transduction pathways. This idea implies that the Drosophila tumor suppressor protein Dlg-A and perhaps its mammalian homolog DLG transmit growth-inhibitory signals from sites of cell–cell contact to downstream effectors. In cells, DLG binds to the free C-terminal end of the tumor suppressor protein APC (11). Considering that APC sustains C-terminal truncations in most sporadic and familial colon cancers (12), it is conceivable that DLG:APC complexes participate in blocking cell cycle progression. If so, one intriguing possibility is that, by binding to DLG, the 9ORF1, HTLV-1 Tax, and HPV E6 oncoproteins prevent the formation of DLG:APC complexes and, thereby, contribute to the promotion of unregulated cellular proliferation. Consistent with this proposal, we found that the 9ORF1 and APC proteins interact preferentially with the same PDZ domain of DLG. Determining the functional consequences of physical interactions between human virus oncoproteins and DLG should help clarify the role of this cellular factor in controlling normal cell growth and in oncogenesis.
Acknowledgments
We thank J. Butel and A. Rice for helpful discussions and J. Butel, L. Donehower, and D. Medina for critical reading of the manuscript. R.S.W. was the recipient of National Science Foundation and U.S. Army Breast Cancer Training Grant (DAMD17-94-J4204) predoctoral fellowships. This work was also supported by National Institutes of Health (CA58541) and American Cancer Society (RPG-97-068-01-VM) grants (R.T.J.).
ABBREVIATIONS
- 9ORF1
Ad9 E4 region ORF 1
- GST
glutathione S-transferase
- DLG
mammalian homolog of Drosophila discs large tumor suppressor protein
- APC
adenomatous polyposis coli protein
- HTLV-1
human T cell leukemia virus type 1
- HPV
human papillomavirus
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF000168).
References
- 1.Horwitz M S. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2149–2171. [Google Scholar]
- 2.Javier R, Raska K, Jr, Macdonald G J, Shenk T. J Virol. 1991;65:3192–3202. doi: 10.1128/jvi.65.6.3192-3202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shenk T. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2111–2148. [Google Scholar]
- 4.Javier R, Raska K, Jr, Shenk T. Science. 1992;257:1267–1271. doi: 10.1126/science.1519063. [DOI] [PubMed] [Google Scholar]
- 5.Javier R T. J Virol. 1994;68:3917–3924. doi: 10.1128/jvi.68.6.3917-3924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss R S, McArthur M J, Javier R T. J Virol. 1996;70:862–872. doi: 10.1128/jvi.70.2.862-872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss R S, Gold M O, Vogel H, Javier R T. J Virol. 1997;71:4385–4394. doi: 10.1128/jvi.71.6.4385-4394.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller B M, Kistner U, Veh R W, Cases-Langhoff C, Becker B, Gundelfinger E D, Garner C C. J Neurosci. 1995;15:2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lue R A, Marfatia S M, Branton D, Chishti A H. Proc Natl Acad Sci USA. 1994;91:9818–9822. doi: 10.1073/pnas.91.21.9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods D F, Bryant P J. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 11.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg G H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 12.Polakis P. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 13.Smith D B, Corcoran L M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. New York: Wiley; 1994. pp. 16.7.1–16.7.7. [Google Scholar]
- 14.Weiss R S, Lee S S, Prasad B V V, Javier R T. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaelin Jr W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 16.Kim S K. Curr Opin Cell Biol. 1995;7:641–649. doi: 10.1016/0955-0674(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 17.Sheng M. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 18.Songyang Z, Fanning A S, Fu C, Xu J, Marfatia S M, Chishti A H, Crompton A, Chan A C, Anderson J M, Cantley L C. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 19.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 20.Kim E, Niethammer M, Rothschild A J, Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 21.Woods D F, Hough C, Peel D, Callaini G, Bryant P J. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevins J R, Vogt P K. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 1. Philadelphia: Lippincott; 1996. pp. 301–343. [Google Scholar]
- 23.Cann A J, Chen I S Y. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 1849–1880. [Google Scholar]
- 24.Howley P M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2045–2076. [Google Scholar]
- 25.Shah K V, Howley P M. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2077–2109. [Google Scholar]
- 26.Chen J J, Reid C E, Band V, Androphy E J. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 27.Desbois C, Rousset R, Bantignies F, Jalinot P. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 28.Kwok R P, Laurance M E, Lundblad J R, Goldman P S, Shih H, Connor L M, Marriott S J, Goodman R H. Nature (London) 1996;380:642–646. doi: 10.1038/380642a0. [DOI] [PubMed] [Google Scholar]
- 29.Yamaoka S, Inoue H, Sakurai M, Sugiyama T, Hazama M, Yamada T, Hatanaka M. EMBO J. 1996;15:873–887. [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner S, Green M R. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 31.Smith M R, Greene W C. J Clin Invest. 1991;88:1038–1042. doi: 10.1172/JCI115364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semmes O J, Majone F, Cantemir C, Turchetto L, Hjelle B, Jeang K T. Virology. 1996;217:373–379. doi: 10.1006/viro.1996.0126. [DOI] [PubMed] [Google Scholar]
- 33.Ponting C P, Phillips C. Trends Biochem Sci. 1995;20:102–103. doi: 10.1016/s0968-0004(00)88973-2. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, Cho K O, Rothschild A, Sheng M. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]