Summary
Transplantation studies and cell lineage analyses require the ability to explicitly distinguish morphologically identical cells that have an identifiable marker indicating their origin in vivo. Several reporter mouse strains have been generated for such studies, but pan-cellular detection of the marker in all tissues has not been achieved. In this report, we describe the generation of transgenic mice that express enhanced green fluorescent protein (EGFP) under control of a 187 kb bacterial artificial chromosome (BAC) containing the murine ROSA26 locus, and show several advantages over existing EGFP reporter lines. It is demonstrated that EGFP is ubiquitously and reproducibly expressed from the murine BAC transgene in all organs and tissues analyzed, including the hematolymphoid compartment. Using this new reporter strain in hematopoietic cell transplantation studies, it is demonstrated that leukocytes in recipients maintain uniform transgene expression and are easily distinguished by flow cytometric analysis of live cells. The results suggest that the ROSA26 BAC is an efficient strategy for expressing complex transgene cassettes in vivo.
Keywords: transgenic, ROSA26, EGFP, hematolymphoid cells, transplantation, bone marrow transplantation
Introduction
The introduction of a genetic marker into transgenic mice represents a powerful approach for analysis of cell lineage and for studying transplantation, cellular differentiation, and organogenesis. Mouse lines that express a marker gene in a restricted number of tissues or cells within a given tissue, or that do not maintain transgene expression regardless of the eventual fate of a given cell, can lead to biased results. Hence, mouse strains that express a marker gene in all tissues and cells throughout development are highly desirable. Several short promoters potentially capable of driving widespread tissue expression of marker genes include SV40 (Furth et al., 1991; Takeda and Toyoda, 1991), CMV (Furth et al., 1991), chicken β-actin (Sands et al., 1993), as well as a combination of CMV and β-actin (Okabe et al., 1997). Although most of these reporter lines show transgene expression in many tissues, the transgene is often not universally and reproducibly expressed within tissues and between animals. This phenomenon of variegated reporter expression may be attributed to position effects of transgene integration into the genome, since small foreign DNA fragments are susceptible to surrounding regulatory elements (Houdebine, 2002).
Production of transgenic mice using bacterial artificial chromosomes (BACs) has become an attractive approach for studying gene expression in vivo. The size of BACs (>100 kb) ensures a high likelihood that a transgene will contain all positive and negative regulatory elements that normally control expression of that gene. In addition, transgenes containing large regions of 5′ and 3′ flanking sequence help to minimize contributions from surrounding genomic elements at the integration site that might alter expression (Houdebine, 2002). The ease and efficiency of introducing desired sequences and deletions into precise sites within BACs by homologous recombination in Escherichia coli makes it possible to generate new reporter lines of mice relatively quickly compared with gene targeting in ES cells (Cotta-De-Almeida et al., 2003; Yu et al., 2000).
The ROSA26 locus, originally identified by gene trapping experiments, directs generalized expression in mice (Friedrich and Soriano, 1991; Zambrowicz et al., 1997). Several reporter genes have been introduced into this locus by targeting in ES cells. However, low levels of expression have limited their utility (Mao et al., 2001; Srinivas et al., 2001). Here, we introduced cDNA sequences encoding enhanced green fluorescent protein (EGFP) into a murine BAC clone containing the ROSA26 locus to generate an indicator mouse line that expresses EGFP at readily detectable levels in all tissues. We directly visualized uniform EGFP fluorescence in all tissues examined, including the hematolymphoid compartment, in both embryos and adult mice. These results demonstrate that the BAC ROSA26-EGFP transgenic line is an effective and efficient mouse model for transplantation and cell fate studies.
Results
Generation of ROSA26-EGFP BAC Transgenic Mice
To determine whether a BAC transgene containing the ROSA26 locus could recapitulate the generalized expression observed with the endogenous gene, we introduced an EGFP cDNA into a 187-kb BAC containing 153 kb of 5′ flanking sequence and 15 kb of 3′ flanking sequence (Fig. 1a) of the ROSA26 gene by homologous recombination in E. coli (Cotta-De-Almeida et al., 2003; Datsenko and Wanner, 2000; Yu et al., 2000). The EGFP cDNA was inserted into the first exon of transcript 1. Two independent transgenic lines were established, each containing a single copy of the transgene. The pattern of EGFP expression in each of the two lines appeared to be identical in each tissue examined. ROSA26-EGFP transgenic mice were born at the expected Mendelian frequency and displayed no overt phenotype, with normal development and fertility.
FIG. 1.

Production of ROSA26-EGFP transgenic mice. (a) The ROSA26-EGFP transgene was generated by introducing EGFP into a BAC containing −153 kb of 5′ flanking sequence and 13 kb of 3′ flanking sequence by homologous recombination in E. coli. (b) Widespread EGFP fluorescence in an e13.5 ROSA26-EGFP heterozygote (left) compared with a wild type control embryo. (c) EGFP expression detected by immunoperoxidase staining in sagittal sections of e13.5 ROSA26-EGFP transgenic (left) and wild-type (right) embryos. Magnification = ×1.25. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The ROSA26 BAC Directs Generalized Expression of EGFP in Mice
Examination of transgenic mice at embryonic day 13.5 (e13.5) showed widespread native EGFP fluorescence in all organs visualized in whole mount preparations (Fig. 1b). Pancellular expression of endogenous EGFP was directly observed in embryonic tissue at higher magnifications of 100× (not shown). Immunohistochemical staining for EGFP confirmed generalized expression of EGFP throughout the embryo at this stage of development (Fig. 1c). These observations suggest that the BAC ROSA26-EGFP transgene appeared to recapitulate the pattern of β-galactosidase expression profile noted in embryos of the original ROSA β-geo26 reporter mouse line (Friedrich and Soriano, 1991; Zambrowicz et al., 1997).
Examination of multiple organs in adult mice for EGFP fluorescence from both pedigrees revealed readily detectable and homogeneous expression in spleen, lung, brain, small intestine, colon, pancreas, testis, kidney, skeletal muscle, liver, heart, and stomach (Figs. 2 and 3). In most tissues, direct EGFP fluorescence colocalized with immunofluorescent staining with an EGFP antibody (not shown). We compared EGFP expression in ROSA26-EGFP mice with that in CAG-EGFP mice, a line frequently used as an EGFP-expressing donor line for transplantation studies, which expresses EGFP under control of the chicken β-actin promoter and cytomegalovirus enhancer (Fukuhara et al., 2005; Hagiwara et al., 2006; Kakui et al., 2005; Okabe et al., 1997). CAG-EGFP mice displayed highly variegated expression of EGFP in the antrum, colon, heart, liver, spleen, and other tissues (Fig. 3). In many tissues from CAG-EGFP mice, some areas produced intense EGFP fluorescence, sometimes stronger than the ROSA26-EGFP mice, while adjacent sections of the same tissue showed minimal or undetectable EGFP expression (Fig. 3). Whereas the ROSA26-EGFP mice showed uniform EGFP fluorescence throughout the alimentary tract, some gastric glands and crypt-villus units of CAG-EGFP mice showed minimal expression but were adjacent to others with intense fluorescence. The fluorescence pattern observed in the GI tract of the CAG-EGFP mouse suggests that the variegated expression may arise from clonal differences occurring in pluripotent cells in the gastric glands and crypt-villus units. The failure to detect uniform EGFP expression in different tissues did not result from tissue damage in the sections as tissue integrity appeared normal based on recognizable structures, DAPI staining of nonexpressing areas, and normal architecture on brightfield examination under Hoffman optics, confirming that the observed EGFP fluorescence in CAG-EGFP mice arose due to variegated expression.
FIG. 2.
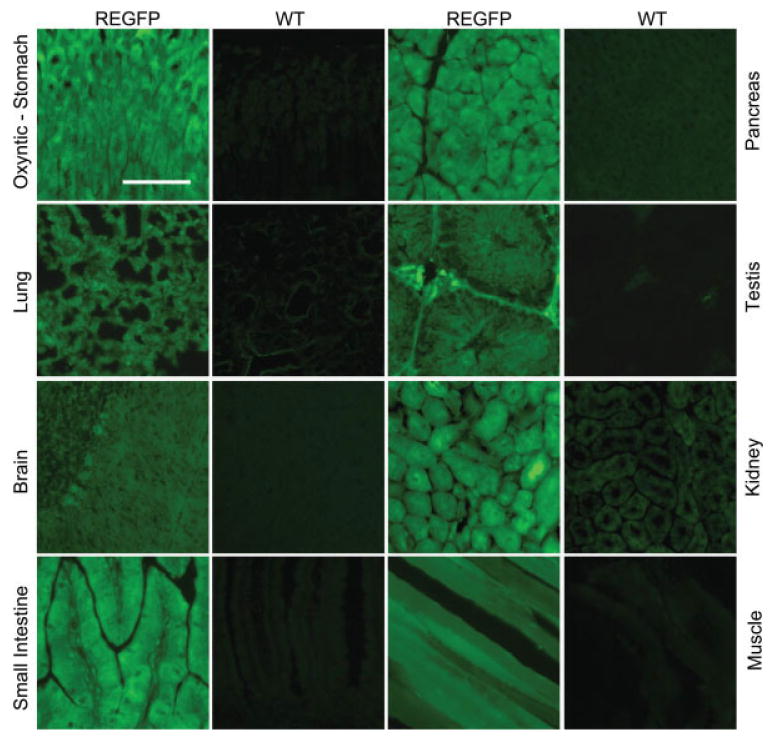
Generalized pancellular expression of EGFP in adult tissues of ROSA26-EGFP (REGFP) transgenic mice. Sections from various organs were examined for native EGFP fluorescence from 6-week-old ROSA26-EGFP and compared with wild-type (WT) littermate controls. Scale bar = 100 µm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
FIG. 3.

Comparison of EGFP expression in tissues from ROSA26-EGFP and the CAG-EGFP mice. ROSA26-EGFP mice show generalized pancellular EGFP expression in sections of various organs at 6 weeks of age, top row. Variegated EGFP fluorescence is seen in CAG-EGFP mouse, middle row. Cells that do not express EGFP are demonstrated with DAPI staining (bottom row) of the same sections shown in the middle row. Scale bar = 100 µm.
Stable and Uniform EGFP Expression from Long Term Repopulating Hematopoietic Stem Cells from ROSA26-EGFP Donors
Transplantation experiments frequently rely upon the ability to identify donor cells expressing a recognizable marker in the recipient mouse to distinguish them from the recipient's cells. The generalized expression of EGFP we observed in the ROSA26-EGFP mouse prompted us to compare its suitability with CAG-EGFP mice as a donor strain for bone marrow transplanted into lethally irradiated mice. Recipient mice were analyzed for EGFP expression at 1, 2, and 4 months posttransplantation to monitor production of myeloid and lymphoid cells from both short- and long-term repopulating stem cells.
At 4 weeks posttransplantation, greater than 90% of B220+ splenocytes, CD4+ and CD8+ thymocytes, and CD11b+ bone marrow myeloid cells examined from ROSA26-EGFP recipients showed bright and uniform EGFP expression as compared with recipients of non-transgenic bone marrow controls. In contrast, recipients of CAG-EGFP bone marrow showed variable EGFP expression at 4 weeks, with ∼90% of B220+ splenocytes but only 50% of CD4+ and CD8+ thymocytes and 65% of CD11b+ bone marrow cells positive for EGFP (data not shown). To assess EGFP expression from long-term repopulating hematopoietic stem cells, we examined hematolymphoid cells from recipients of ROSA26-EGFP and CAG-EGFP donor marrow at 4 months posttransplantation by flow cytometry (Fig. 4). Most CD11b+ myeloid cells (95%) in the bone marrow of ROSA26-EGFP recipients showed relatively uniform EGFP expression, with mean fluorescence intensity greater than 100 fold that of nontransplanted mice. In contrast, only a fraction (38%) of bone marrow CD11b+ cells in recipients of CAG-EGFP marrow expressed EGFP at mean levels only 10 fold higher than nontransplanted mice, with a broad distribution of fluorescence intensity over a several thousand-fold range. Likewise, in peripheral blood, most CD11b+ myeloid cells (monocytes and neutrophils) in recipients of ROSA26-EGFP marrow exhibited bright EGFP fluorescence, whereas the majority of circulating CD11b+ leukocytes in mice transplanted with CAG-EGFP marrow did not express EGFP, with the remainder expressing EGFP over a broad fluorescence range.
FIG. 4.
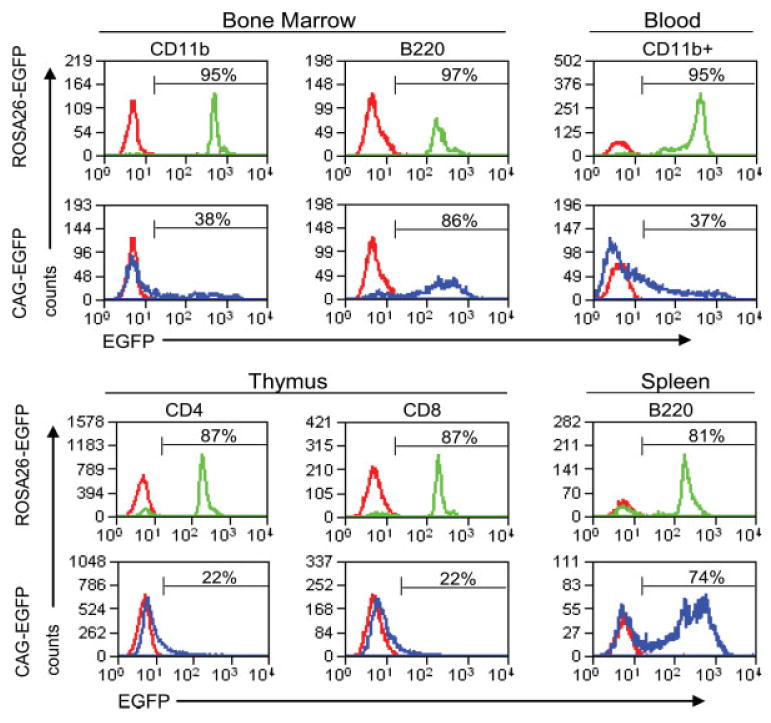
EGFP expression in hematolymphoid cells from mice transplanted with ROSA26-EGFP or CAG-EGFP bone marrow, analyzed 4 months posttransplantation. EGFP expression in leukocytes from the indicated tissues was assessed in bone marrow and peripheral blood (top panels) and thymus and spleen (bottom panels) of recipients of bone marrow from ROSA26-EGFP transgenic donors (green lines), CAG-EGFP transgenic donors (blue lines), and nontransgenic syngeneic donors (red lines). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Lymphocytes repopulating irradiated mice transplanted from ROSA26-EGFP donors showed much greater uniformity and percentage of cells expressing detectable EGFP than their counterparts from recipients of marrow from CAG-EGFP mice. In particular, 87% of CD4+ and CD8+ thymocytes strongly expressed EGFP in the ROSA26-EGFP recipients, whereas only a minor fraction (<25%) of CD4+ or CD8+ thymocytes were positive for EGFP expression in CAG-EGFP recipients (Fig. 4). Although we did not perform simultaneous staining for both CD4 and CD8, it is likely that both single- and double-positive thymocytes expressed EGFP in ROSA26-EGFP recipients. A high percentage of splenic and circulating B cells (B220+) in recipients of either strain expressed EGFP, although the recipients of ROSA26-EGFP donor marrow showed a somewhat higher percentage with increased uniformity compared with animals receiving CAG-EGFP bone marrow.
Discussion
Transgenic mice that uniformly express readily detectable markers in all cells of all tissues are an essential tool for marking cells for transplantation studies, whole animal chimera experiments, and cell lineage analyses. Several lines of mice created for this purpose partially fulfill the criteria of pancellular expression in some but not all tissues, making it potentially difficult to identify the origin of all cells in transplant recipients. We have generated ROSA26EGFP transgenic mouse lines expressing EGFP under control of a 187-kb BAC containing the murine ROSA26 locus that have several advantages over existing models. In our experience, using homologous recombination in Escherichia coli is an efficient and rapid approach for generating transgenic mice, as it enables generation of BAC transgenes ready for pronuclear microinjection in 3 weeks. The identified murine BAC appears to contain sequences sufficient to recapitulate the generalized expression directed by the endogenous ROSA26 locus as originally reported (Friedrich and Soriano, 1991; Zambrowicz et al., 1997).
The ROSA26-EGFP transgene is efficiently and uniformly expressed in the differentiated progeny of transplanted HSCs, making their identification unambiguous, and this expression is much more faithful than that conferred by HSCs from CAG-EGFP donors. It is difficult or impossible to establish the origin of leukocytes in recipients of CAG-EGFP hematopoietic stem cells, owing to the much lower percentage of cells expressing EGFP above the background level. Only a fraction of the major lymphoid lineages were EGFP positive in the recipients of CAG-EGFP bone marrow, which is consistent with the variegated EGFP expression in splenic tissue of CAG-EGFP mice. The presence of high percentages of CD11b+ EGFP expressing cells 4 months after engraftment in ROSA26-EGFP recipients suggests that EGFP is expressed in long-term hematopoietic stem cells that give rise to a relatively uniform population of descendant lineages (Lagasse and Weissman, 1994; Morrison and Weissman, 1994; Wright et al., 2001).
In contrast to recipients of CAG-EGFP mice, most (95%) myeloid cells in recipients of ROSA26-EGFP mice showed at least 10- to 100-fold more EGFP fluorescence than control donor cells. Analysis of donor ROSA26-EGFP lymphocytes also showed a small fraction of EGFP− cells (Supplemental Fig. 1). For each of the lineages examined, the fraction of EGFP+ cells was slightly higher in donor mice than the BMT recipients. Likewise, a much higher percentage of CAG-EGFP lymphocytes were EGFP− compared with the ROSA26-EGFP mice (38% vs. 14%, not shown). Thus the small fraction of lymphoid and myeloid cells in ROSA26-EGFP recipients showing a lower level EGFP expression may result from a combination of factors including low level variegated expression of EGFP, rare cells that survived irradiation, rare nondonor cells from the recipient that bind to lineage specific antibodies used for FACS, or a moderate engraftment defect related to histoincompatibility. Although the ROSA26-EGFP donors were not fully backcrossed to the recipient background, any hybrid resistance would favor engraftment and stable expression by the CAG-EGFP donor cells, which are congenic with their recipient. Therefore, the ROSA26-EGFP mice are superior donors for any experiments that track hematopoietic cell fate in transplant recipients.
The inability to achieve the desired pattern of expression for a given transgene is a common occurrence. In some cases, relatively short transgenes failed to include distant yet essential transcription regulatory sequences required for cell-type specific expression, leading to either misexpression in inappropriate cells or loss of expected expression in other cells. A second problem encountered with short transgenes is their susceptibility to the effects of chromosomal integration, where enhancers or silencers in neighboring genes modify expression (Ogbourne and Antalis, 1998). Integration site effects often lead to marked differences in expression patterns between independent pedigrees because of insufficient sequence to insulate the transgene from effects of nearby genes. Position effect variegation, with variable rather than uniform expression in similar cells within a single tissue, represents another effect of integration site on expression patterns. The commonly used CAG-EGFP mouse, which expressed EGFP in a short transgene, exhibits extensive variegation. Another transgene containing only 0.8 kb of the ROSA26 locus (Kisseberth et al., 1999) directed widespread EGFP expression that was uniform in a number of tissues. However, some tissues, including liver and intestine, showed considerable position effect variegation in these mice.
Insertion of a reporter into the ROSA26 locus by homologous recombination in ES cells to generate a “knock in” mouse is one approach that ensures widespread expression because of the presence of the native chromosomal sequences extending millions of bp flanking the locus. Several groups have knocked-in genes expressing fluorescent proteins, such as EGFP, EYFP, and ECFP, into the ROSA26 locus to improve the general utility of this reporter line (Mao et al., 2001; Srinivas et al., 2001). Anecdotal reports suggest that knocking EGFP into endogenous loci frequently results in insufficient EGFP expression for direct visualization in tissue sections (Gong et al., 2003). A single copy of the ROSA26-EGFP BAC transgene universally expresses EGFP at levels allowing visualization of endogenous EGFP fluorescence in organ whole mounts as well as in thin tissue sections and live cells. The ROSA26-EGFP transgenic mice described here show an approximately 100-fold increase in EGFP fluorescence in lymphocytes compared with nontransgenic mice with minimal overlap in intensity between the two. FACS analysis of EGFP expression in lymphocytes from a mouse with EGFP knocked-in to the ROSA26 locus showed a less than one log increase in fluorescence with substantial overlap between transgenic and nontransgenic lines (Mao et al., 2001). These results suggest that EGFP expression in lymphocytes of the ROSA26-EGFP BAC mice is considerably higher and easier to distinguish from background than lymphocytes from the previously described “knock in” mice (Mao et al., 2001). The high level of EGFP fluorescence seen with both pedigrees, compared with the knock-in, suggests that distant inhibitory elements in the ROSA26 locus that reduce transcription of endogenous ROSA26 gene were not included in the transgene. The loss of these regulatory elements appears to enhance the expression of EGFP while enough sequence is still present in the BAC to accurately reproduce the generalized expression of the endogenous gene (Friedrich and Soriano, 1991).
In addition to the use of the ROSA26 locus as a reporter line for transplantation and cell lineage analysis, mice with conditional ROSA26 alleles represent a powerful approach to activate expression of any gene in specific cell types at different stages of development. A gene inserted into the ROSA26 BAC with upstream floxed transcription termination sequences can be expressed as desired with the appropriate Cre deleter strain. Rapid production of transgenes in the ROSA26 BAC by recombineering as described here is potentially a new, highly efficient approach for generating lines of mice expressing a variety of proteins in any tissue desired.
Methods
BAC Transgene Construction and Purification
A murine BAC clone RP23-401D9 containing the ROSA26 locus was obtained from ResGen Invitrogen. The red-shifted variant EGFP cDNA (BD Biosciences Clonetech No. 6029-1) containing simian virus 40 polyadenylation signals 3′ to the coding sequence was introduced into a BAC containing the ROSA26 locus by homologous recombination in E. coli as previously described (Schonhoff et al., 2004). A linear fragment containing the amplified EGFP cassette was introduced at nucleotide position 521 (see GenBank accession no. U83173) of exon 1 of transcript 1 of the ROSA26 gene (Zambrowicz et al., 1997) (PCR primers: sense = GGGACTCTGGCGGGAGGGCGGCTTGGTGCGTTTGCGGGGATGGTGAGCAAGGGCGAGGAGCTGT; antisense = ACGGCTCCACCACGCTCGGAGGGCCTGCCGCGGCCGCCCATCCTCCTTAGTTCCTATTCCGA, the italicized sequence is homologous to the sequence flanking the ROSA26 translation start site). As was originally described, tens to hundreds of BAC containing colonies grew after antibiotic selection (Datsenko and Wanner, 2000). Most of the selected colonies contained the desired insertion and flanking BAC sequences when examined by Field Inversion Gel Electrophoresis and DNA sequencing. Modified BAC DNA was purified for pronuclear injection using the NucleobondAX (BD Bioscience) alkaline lysis-based procedure and resuspended in injection buffer (10 mM Tris-HCL pH 7.5, 0.1 mM EDTA, 30 µM spermine, 70 µM spermidine, 100 mM NaCl).
Production of Transgenic Mice
Purified circular ROSA26-EGFP BAC DNA (1 ng/µl) was microinjected into pronuclei of fertilized oocytes of B6XB6D2F1 mice. Founder mice were genotyped by DNA amplification using primers specific for the EGFP cassette (PCR primers: sense = 5′ CCCTGAAGTTCATCTGCA, antisense = 5′ AGAGCGTTCACCGACAAA). Two independent transgenic lines were identified and pedigrees were maintained on a CD1 background. Transgene copy number was determined by quantitative PCR using primer pairs specific to EGFP (PCR primers: sense = 5′ CCCTGAAGTTCATCTGCA, antisense = 5′ CCCCAGGATGTTGCCGTCC).
Histology and Immunohistochemistry
Adult tissues and embryos were fixed with 4% paraformaldehyde for 6 h on ice, washed with phosphate-buffered saline, and equilibrated with 30% sucrose phosphate-buffered saline at 4°C. Frozen sagittal sections (12 µm) were imaged directly for EGFP fluorescence or immunostained with Rabbit anti-EGFP (Abcam) 1:10,000.
Bone Marrow Transplantation
Bone marrow was harvested from donor transgenic animals according to standard methods. CD1 recipient mice (6–8 weeks old, n = 10) were lethally irradiated (1,000 cGy) and intravenously injected via the tail vein with 2 × 106 whole bone marrow cells isolated from ROSA26-EGFP transgenic donor mice, backcrossed two generations from D2B6 to CD1. C57Bl/6 recipients (6–8 weeks old, n = 10) were lethally irradiated (1,150 cGy) and transplanted with 2 × 106 CAG-EGFP transgenic bone marrow cells isolated from congenic C57Bl/6 donors. Engraftment of bone marrow in recipient mice was determined by analysis of EGFP expression in leukocytes from peripheral blood, bone marrow, spleen, and thymus at 1, 2, and 4 months posttransplantation. Engrafted recipients appeared healthy at the time of analysis and showed no evidence of infection or graft-versus-host disease. Single cell suspensions were generated from each tissue, stained with R-Phycoerythrin-conjugated antibodies for CD4, CD8, Cd11b, and B220, and analyzed by flow cytometry for EGFP and PE on a Dako Cyan flow cytometry analyzer, using Cytomation Summit software.
Supplementary Material
This article contains Supplementary Material available via the Internet at http://www.interscience.wiley.com/jpages/1526-954X/suppmat.
Acknowledgments
The authors acknowledge the assistance of the Tufts Transgenic Core Facility.
Contract grant sponsor: NIH, Contract grant numbers: DK43673, DK52870, DK67166, CA90576, CA105043; Contract grant sponsor: GRASP Digestive Disease Center, Contract grant numbers: P30-DK34928, T32-DK007542
Literature Cited
- Cotta-De-Almeida V, Schonhoff S, Shibata T, Leiter A, Snapper SB. A new method for rapidly generating gene-targeting vectors by engineering BACs through homologous recombination in bacteria. Genome Res. 2003;13:2190–2194. doi: 10.1101/gr.1356503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Tomita S, Nakatani T, Yutani C, Kitamura S. Endogenous bone-marrow-derived stem cells contribute only a small proportion of regenerated myocardium in the acute infarction model. J Heart Lung Transplant. 2005;24:67–72. doi: 10.1016/j.healun.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Furth PA, Hennighausen L, Baker C, Beatty B, Woychick R. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 1991;19:6205–6208. doi: 10.1093/nar/19.22.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hagiwara H, Ohsawa Y, Asakura S, Murakami T, Teshima T, Sunada Y. Bone marrow transplantation improves outcome in a mouse model of congenital muscular dystrophy. FEBS Lett. 2006;580:4463–4468. doi: 10.1016/j.febslet.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Houdebine LM. The methods to generate transgenic animals and to control transgene expression. J Biotechnol. 2002;98:145–160. doi: 10.1016/s0168-1656(02)00129-3. [DOI] [PubMed] [Google Scholar]
- Kakui K, Itoh H, Sagawa N, Yura S, Takemura M, Kawamura M, Fujii S. Experimental transplantation study for possible transformation of bone marrow cells in the mouse placenta. Placenta. 2005;26:678–685. doi: 10.1016/j.placenta.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Kisseberth WC, Brettingen NT, Lohse JK, Sandgren EP. Ubiquitous expression of marker transgenes in mice and rats. Dev Biol. 1999;214:128–138. doi: 10.1006/dbio.1999.9417. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Weissman IL. bcl-2 inhibits apoptosis of neutrophils but not their engulfment by macrophages. J Exp Med. 1994;179:1047–1052. doi: 10.1084/jem.179.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Ogbourne S, Antalis TM. Transcriptional control and the role of silencers in transcriptional regulation in eukaryotes. Biochem J. 1998;331(Part 1):1–14. doi: 10.1042/bj3310001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Sands AT, Hansen TN, Demayo FJ, Stanley LA, Xin L, Schwartz RJ. Cytoplasmic β-actin promoter produces germ cell and preimplantation embryonic transgene expression. Mol Reprod Dev. 1993;34:117–126. doi: 10.1002/mrd.1080340202. [DOI] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Toyoda Y. Expression of SV40-lacZ gene in mouse pre-implantation embryos after pronuclear microinjection. Mol Reprod Dev. 1991;30:90–94. doi: 10.1002/mrd.1080300203. [DOI] [PubMed] [Google Scholar]
- Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA β geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This article contains Supplementary Material available via the Internet at http://www.interscience.wiley.com/jpages/1526-954X/suppmat.


