Abstract
Understanding the molecular architectures of enveloped and complex viruses is a challenging frontier in structural biology because in most cases, the structural and compositional variation from one viral particle to another precludes the use of either crystallization or image averaging procedures that have been successfully implemented in the past for highly symmetric viruses. While advances in cryo electron tomography of unstained specimens provide new opportunities for identification and molecular averaging of individual subcomponents such as the surface glycoprotein spikes on these viruses, electron tomography of stained and plunge-frozen cells is being used to visualize the cellular context of viral entry and replication. Here, we review recent developments in both areas as they relate to our understanding of the biology of heterogeneous and pleiomorphic viruses.
Introduction
Advances over the last several decades in biological electron microscopy and X-ray crystallography have led to fascinating insights into the architectures of a wide range of viruses with diameters ranging from ~30 nm to ~300 nm. Knowledge of the arrangement of the internal viral components such as the capsid representing the packaged nucleic acid and the exterior, generally composed of proteins that mediate entry into cells has been vital not only for improved understanding of the principles of viral organization, but also for the development of strategies to combat viral infection leading to a variety of human diseases. Early structural analyses using electron microscopy were largely devoted to obtaining projection images of viruses in infected cells that were fixed, stained with electron-dense reagents and resin-embedded for analysis using electron microscopy at room temperature. Over the last decade, the advent of cryo electron microscopic approaches has provided new opportunities to investigate the structures of intact viruses without staining, but by analysis of plunge-frozen specimens [1]. Combined with image averaging methods, especially for viruses displaying high levels of internal symmetry, these methods have led to descriptions of viral architecture at sub-nanometer resolutions, and interpretation of subunit arrangements at atomic resolution using structures of individual components determined by X-ray crystallography [2,3].
Until recently, three-dimensional electron microscopic analysis of the native states of viruses without intrinsic symmetry such as retroviruses and other enveloped viruses has remained largely unexplored. However, the emergence of powerful technologies for carrying out electron tomography of viruses in isolation and in infected cells is rapidly beginning to provide novel glimpses of viral architecture in enveloped and complex viruses [4–10]. Here, we review some recent areas of progress and the challenges that lie ahead in this field.
Tomography vs. single particle reconstruction methods
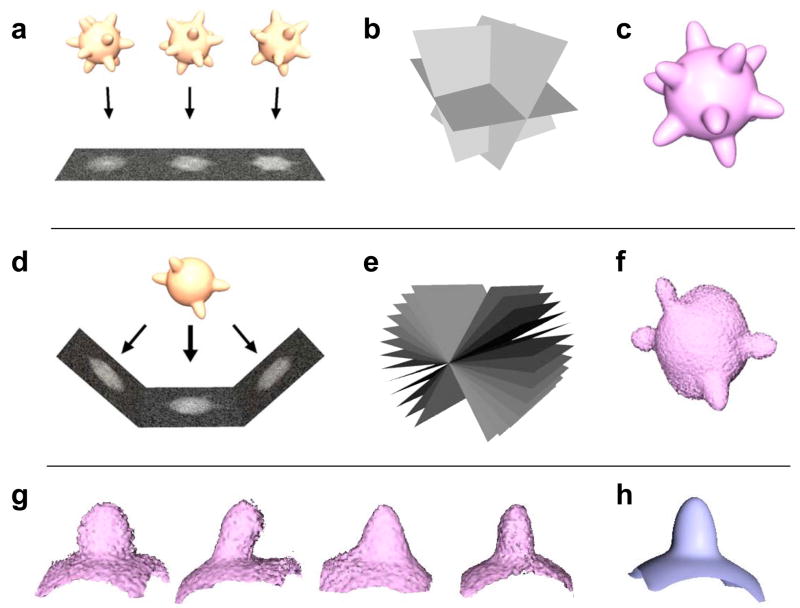
Approaches to derive the virus structures of 3D objects by single particle electron microscopy are based on the fundamental premise that projection images of different copies of the same virus can be averaged together. The strategy for reconstructing 3D structure from a series of projection images takes advantage of the fact that the 2D Fourier transform of a projection image corresponds to a central slice through the origin of the 3D Fourier transform of the object; thus, as the 3D Fourier transform is progressively built up by including more and more projection images, the inverse of the transform provides progressively improved representations of the three-dimensional structure of the object being imaged (figs. 1a–c). Because of the necessity of using low doses to preserve structural integrity during electron microscopic imaging, the presence of symmetry makes this type of averaging extremely effective, and has driven rapid advances in 3D reconstructions of viruses and their components displaying icosahedral symmetry [11–13], and its application to study dynamical aspects of virus structure [14,15].
Figure 1.
Strategies for 3D reconstruction and averaging in 3D electron microscopy. In single particle electron microscopy, projection images of identical copies of the object (a), are averaged together by determining relative orientations (b), of the projection images recorded over a range of orientations and combined to generate a three-dimensional representation of the object (c). In electron tomography, multiple views (d) of the same object are recorded at different orientations relative to the beam (e) and combined using back-projection-based or related methods to generate a three-dimensional representation of the object (f). The distortion in the shape of the object is because the reconstruction is based on views from a limited angular range (+/− 60° in this case) that is experimentally accessible in data collection geometries currently used in electron tomography. The four protrusions on the object in this example are chosen to illustrate individual spikes on the surface of a virus. The 3D volumes of these spikes extracted from the reconstruction in (f) are presented in (g). Despite the presence of distortions rising from the “missing wedge” in the individual recovered volumes, when correctly compensated for, they can be combined to generate an averaged 3D volume. In cryo electron tomography, where low doses are used to preserve specimen integrity, this latter strategy allows the use of molecular averaging methods to determine the structures of subcomponents of viruses such as viral spikes at higher resolution.
For pleiomorphic viruses, where each virion can display a different shape and size, 3D reconstruction by image averaging is not possible, but tomographic analysis can be used to determine the overall 3D structure. In electron tomography [16], a series of projection images is generated by rotating the specimen over a tilt range that is typically restricted to +/− 70° in most cases because of physical limitations of specimen geometry in the electron microscope column, and due to the rapid increase in effective specimen thickness of the flat specimen at the highest tilt angles. These projections can then be combined to generate a three-dimensional representation of the object. In using this strategy to visualize the structures of viruses, there are two fundamental features worthy of note that affect the final outcome. First, the fact that the range of angles sampled is limited results in a distortion of the reconstructed object (figs. 1d–f). Second, the signal-to-noise ratio in the 3D reconstruction is limited by the fact that the electron dose is spread out over the entire tilt series, and results in relatively noisy 3D representations as compared to 3D structures derived by averaging large numbers of viruses using single particle approaches.
There is, however, the exciting prospect that these limitations can be overcome by the use of image averaging approaches as applied to selected regions within tomograms of whole viruses and cells. Multiple copies of potentially homogeneous subcomponents of heterogeneous objects can be selected out, and averaged to obtain the 3D structure of the subcomponent at improved resolution. This idea is schematically illustrated in figs. 1g and 1h where the 3D volumes of the four surface protrusions evident in the object in fig. 1f are extracted (fig. 1g) and averaged together (fig. 1h) to generate a 3D structure derived by coherent averaging of the four extracted subvolumes. The striking difference between the visual appearance of the four extracted subvolumes arises solely from differences in orientation between the principal axis of the protrusion and the direction of imaging, a problem referred to as the “missing wedge” effect. Techniques to average volumes while correctly accounting for this missing wedge and with proper classification to ensure that the entities being averaged belong to the same structure are extremely important, and are being developed in a number of research groups [17–19].
Tomographic analyses of viruses
Application of tomography to investigate the architectures of immunodeficiency viruses, members of the herpesvirus family and influenza virus have begun to yield novel information combined with a good measure of controversy (see fig. 2). Some of these studies, such as those with HSV-1 [6] and influenza virus [20], have involved descriptive studies of viral architecture indicating the nature of structural pleiomorphy found in the assembly of these viruses. In a similar vein, Briggs et al. [21] have reported on the assembly of the HIV-1 core by tomography of formaldehyde-treated, plunge-frozen virions. From inspection of tomograms of several virus particles, they have concluded that core assembly is likely to be initiated at the narrow end, and proceeds towards the distal side until the viral membrane limits its growth. It will be interesting to test whether this type of mechanism will also hold for other retroviruses, and whether this sequence of assembly has some deeper functional significance. In complementary studies, Wright et al. [22] have presented images of immature HIV-1 rendered non-infectious by point mutations in multiple Gag components, and suggest hypothetical models for the relative spatial arrangement of cleaved Gag components in the interior of the virus.
Figure 2.
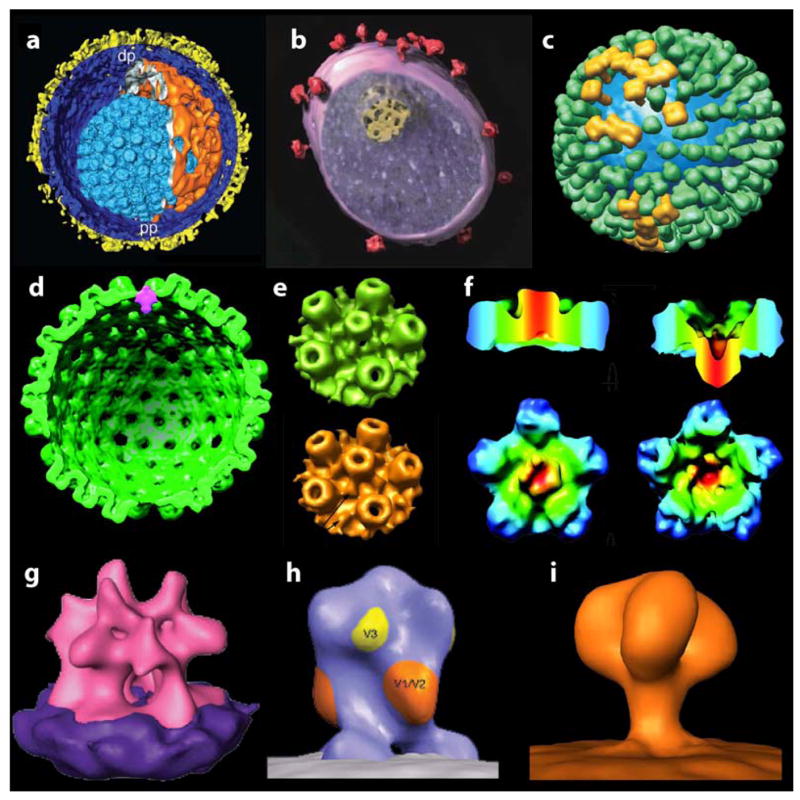
Recent examples of the use of cryo electron tomography to visualize the structures of viruses and their components. The examples presented are segmented renderings of the 3-D structures of (a) herpes simplex virus (HSV) capsid [6], (b) simian immunodeficiency virus [7], (c) influenza virus [20], (d–f) views of the herpesvirus capsid portal vertex from three different groups: a cross-sectional rendering (d) highlighting the unique portal vertex in HSV-1 by Chang et al. [24], rendering of surface views (e) of portal (top) and non-portal vertices (bottom), also of HSV-1 by Cardone et al. [23] and views of the Kaposi’s sarcoma-associated herpesvirus (f) with non-portal penton vertex (left) and portal (right) vertex shown by Deng et al. [25], (g) averaged structure of the surface spike from moloney murine leukemia virus [5] and (h,i) divergent, averaged 3D structures of the surface spike on SIV derived from the work of Zhu et al. [10] and Zanetti et al. [9].
Investigation of the entry portal in herpesvirus capsids by three different groups [23–25] provides another glimpse of the use of tomography in analysis of viral structure. There are many similarities in structure between principles of capsid assembly in tailed bacteriophages [26] and herpesviruses, leading to the question of how one of the twelve 5-fold vertices of the capsid shell becomes uniquely assigned to the entry portal. Tomographic studies reported by all three groups verify the existence of this portal at a unique vertex; however there is disagreement about the precise position of the portal at this vertex. Thus, while Chang et al. [24] and Deng et al. [25] conclude that the bulk of the portal mass lies at a level that corresponds to the floor of the capsid, and points inward, Cardone et al. [23] argue that the portal is mounted pointing outwards from the capsid floor. The discrepancy in these studies may have to do with the use of different capsid preparations, differences in biochemical treatment of the capsids prior to plunge-freezing, and variations in the computational procedures used. Improvements in resolution of the reconstructions can be expected to resolve some of these ambiguities before long.
Two recent reports by Zhu et al. and Zanetti et al. [9,10] on the averaged 3D structures of the surface glycoprotein spike on simian immunodeficiency virus (SIV) have sparked a different kind of controversy. Both groups used very similar virus preparations and obtained what appear to be very similar images, but with very different final outcomes for the structure. The reasons for this discrepancy, and the divergent conclusions about “three- vs. one-legged” viral spikes probably has much to do with differences in the methods used by the different groups for 3D averaging and the errors that arise when proper classification strategies and corrections for the missing wedge have not been employed, as has been pointed out in a commentary on the findings reported in these two papers [8]. Controversy notwithstanding, it is clear that this is only the beginning of efforts to combine cryo electron tomography and molecular averaging methods to obtain penetrating insights into the tertiary and quaternary structures of spikes on these and related viruses.
Virus-antibody complexes and the cellular interface
Understanding how viruses are processed by the host organisms they invade is of central interest from a biological and medical perspective. Historically, these studies have spanned both interactions of viruses with antibodies as well as studies by electron [27–29] and light microscopy [30] of various stages in the viral life cycle. Recent progress in both areas provides encouraging signs that the use of 3D electron microscopic approaches could also be invaluable in the physiological context.
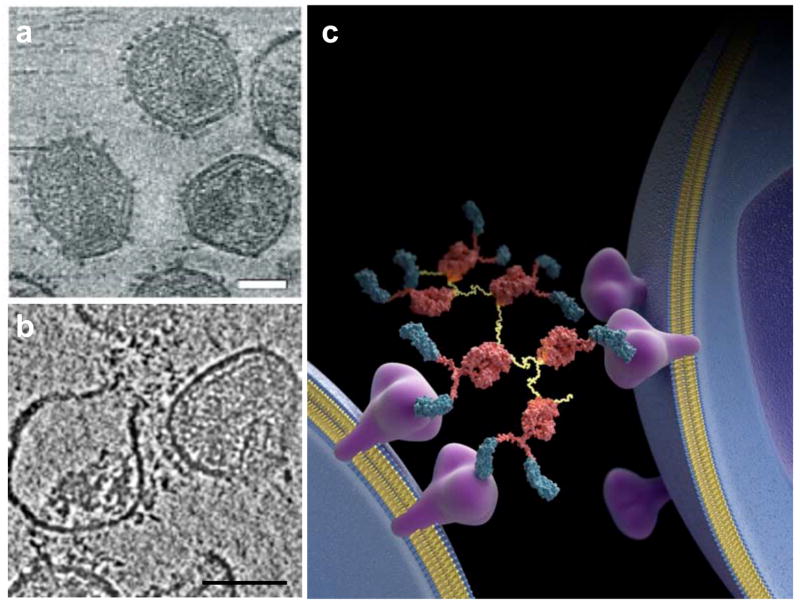
For icosahedral viruses, the use of symmetry has been exploited to localize binding sites of antibodies and antibody fragments on the viral surface [31,32]. In the case of viruses lacking symmetry, understanding these interactions is more challenging. One example where tomography has proved to be useful has been reported in a recent study of the binding of a highly potent neutralizing antibody construct to native gp120 on immunodeficiency viruses. Bennett et al [33] have shown that the potent HIV neutralizing antibody construct D1D2-IgP works by bridging neighboring spikes on the same virus and spikes on neighboring viruses by virtue of its flexible, polyvalent structure, suggesting that spike cross-linking is likely to be central to the mechanism of neutralization by compounds with these characteristics (fig. 3). Intriguingly, the presence of bound neutralizing antibody promoted not only neutralization, but also viral rupture. This observation suggests new physical principles that could be used for design of effective therapeutic candidates for treatment of AIDS by destruction of infectious virions, in contrast to the previous emphasis on simple neutralization of viral spikes.
Figure 3.
Cryo-electron tomography of SIV complexed with the potent neutralizing protein D1D2-IgP [7,33]. (a,b) Tomographic slices from 3D reconstructions of purified SIV in the absence of, (a) and in complex with the potent neutralizing protein D1D2-IgP, which promotes an increase in the frequency of viral rupture leading to loss of internal content of the virus (b). (c) Schematic illustration of the finding that the polyvalent D1D2-IgP antibody can cross-link spikes on, and across viruses. Scale bars in (a) and (b) are 50 nm and 100 nm, respectively.
The extension of virus tomography to study different stages of the viral life cycle is an obvious and important extension of studies on purified viruses. Studies on vaccinia virus in free and cell-associated states have been interpreted to provide evidence for a model in which cell attachment is required to induce decondensation of the viral DNA, and for a mechanism by which viral DNA is released into the cytoplasm by opening of the core on the side [4,34]. Electron tomographic studies on cells infected with flock house virus, a positive strand RNA virus, have been used to localize the 50 nm-sized compartments where viral RNA is synthesized and to delineate the architecture and stoichiometry of the viral replication complex [35]. Recent studies of HIV and SIV entry into CD4+ T-cells provide another example where tomographic analyses of virus-infected cells have led to novel functional insights (fig. 4). Sougrat et al. [7] have shown that docking of SIV or HIV-1 on the T cell surface occurs via a neck-shaped contact region that is ~40 nm wide and is consistently composed of a closely spaced cluster of rod-shaped features, termed the viral “entry claw”. These examples, taken together with reports of the architectures of nascent HSV-1 virions in infected cells [36] bode well for the use of tomography of frozen-hydrated as well as stained, plastic-embedded cells to extend the understanding of virus-cell interactions into the third dimension.
Figure 4.
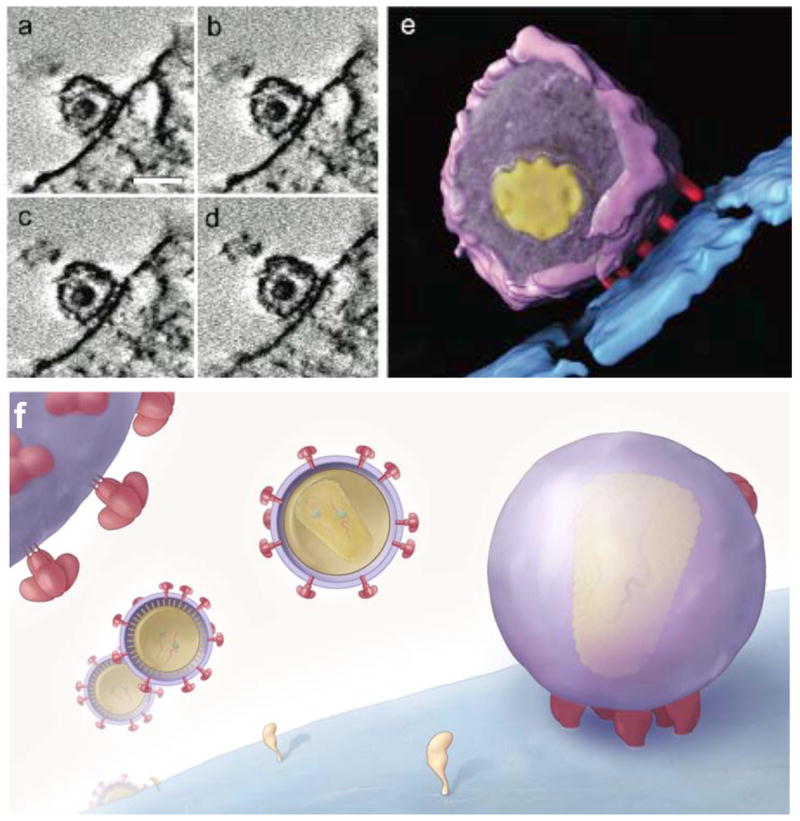
Electron tomographic analysis of the contact between HIV and T-cells. Tomographic slices (a–d) and segmented representation (e) of the distinctive contact zone between HIV-1 and CD4+ T-cells termed the viral “entry claw”, which is not observed in the presence of the peptide entry inhibitor C34, the CCR5 antagonist TAK779, or anti- CD4 antibodies. The schematic in (f) illustrates the general architecture of the ~40 nm wide entry claw, which is composed of about 5–7 rods of density and the fact that viruses engaged in this type of contact appear to have few detectable remaining spikes on their surface in contrast to viruses that have not yet made cellular contact. Scale bars in (a) –(d) are 100 nm long.
Conclusion
Transmission electron microscopy is limited in practice to imaging specimens with thicknesses well below a micron, even with the use of microscopes at the highest operating voltages. Viruses and thin sections of the interior of larger eukaryotic cells meet these physical requirements, and their study using 2D and 3D electron microscopic approaches is providing fascinating insights into the biology of virus assembly, entry and replication. The use of cryo electron tomographic approaches to analyze and quantitate the structural complexity of non-symmetric viruses could be of immediate medical relevance in designing strategies to combat viral diseases. The use of tomography to provide structural information at a resolution intermediate to that obtained by X-ray crystallography and light microscopy thus bridges a critical gap in the biomedical imaging spectrum.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
- 1.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 2.Baker TS, Olson NH, Fuller SD. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cyrklaff M, Risco C, Fernandez JJ, Jimenez MV, Esteban M, Baumeister W, Carrascosa JL. Cryo-electron tomography of vaccinia virus. Proc Natl Acad Sci U S A. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grunewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 7.Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. Report of electron tomographic analysis of early events in the entry of HIV and SIV into T-cells leading to the discovery of the viral “entry claw”, a distinctive structure formed at the contact zone between virus and cell. Understanding the structure of the entry claw may facilitate the identification of improved drugs for the inhibition of HIV-1 entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subramaniam S. The SIV surface spike imaged by electron tomography: one leg or three? PLoS Pathog. 2006;2:e91. doi: 10.1371/journal.ppat.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 11.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 12.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 13.Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 A. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 14.Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10:334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- 15.Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci U S A. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntosh R, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Bartesaghi A, Sprechmann P, Randall G, Sapiro G, Subramaniam S. 2007 doi: 10.1016/j.jsb.2008.02.008. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster F, Pruggnaller S, Seybert A, Frangakis AS. Classification of cryo-electron sub-tomograms using constrained correlation. J Struct Biol. 2007 doi: 10.1016/j.jsb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. Embo J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361:426–434. doi: 10.1016/j.virol.2006.10.047. One of three papers concurrently published papers on structural analysis of the entry portal of herpes simplex virus type 1 using cryo electron tomography. Each of these studies confirms that the structure of the portal is different when compared to the remaining eleven vertices of the viral capsid; however they disagree on the precise architecture of the portal itself. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81:2065–2068. doi: 10.1128/JVI.02053-06. see ref. 23 annotation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng B, O'Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi's sarcoma-associated herpesvirus capsid by cryoelectron tomography. J Virol. 2007;81:3640–3644. doi: 10.1128/JVI.02254-06. see ref. 23 annotation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leiman PG, Kanamaru S, Mesyanzhinov VV, Arisaka F, Rossmann MG. Structure and morphogenesis of bacteriophage T4. Cell Mol Life Sci. 2003;60:2356–2370. doi: 10.1007/s00018-003-3072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orenstein JM. Replication of HIV-1 in vivo and in vitro. Ultrastruct Pathol. 2007;31 :151–167. doi: 10.1080/01913120701344343. [DOI] [PubMed] [Google Scholar]
- 28.Ojeda S, Domi A, Moss B. Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex. J Virol. 2006;80:9822–9830. doi: 10.1128/JVI.00987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy AM, Parker JS, Parrish CR, Whittaker GR. Early stages of influenza virus entry into Mv-1 lung cells: involvement of dynamin. Virology. 2000;267:17–28. doi: 10.1006/viro.1999.0109. [DOI] [PubMed] [Google Scholar]
- 30.Brandenburg B, Lee LY, Lakadamyali M, Rust MJ, Zhuang X, Hogle JM. Imaging Poliovirus Entry in Live Cells. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway JF, Watts NR, Belnap DM, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Characterization of a conformational epitope on hepatitis B virus core antigen and quasiequivalent variations in antibody binding. J Virol. 2003;77 :6466–6473. doi: 10.1128/JVI.77.11.6466-6473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geldmacher A, Skrastina D, Petrovskis I, Borisova G, Berriman JA, Roseman AM, Crowther RA, Fischer J, Musema S, Gelderblom HR, et al. An amino-terminal segment of hantavirus nucleocapsid protein presented on hepatitis B virus core particles induces a strong and highly cross-reactive antibody response in mice. Virology. 2004;323:108–119. doi: 10.1016/j.virol.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 33.Bennett AE, Liu J, Van Ryk D, Bliss D, Arthos J, Henderson RM, Subramaniam S. Cryo electron tomographic analysis of an HIV neutralizing protein and its complex with native viral gp120. J Biol Chem. 2007 doi: 10.1074/jbc.M702025200. [DOI] [PubMed] [Google Scholar]
- 34.Cyrklaff M, Linaroudis A, Boicu M, Chlanda P, Baumeister W, Griffiths G, Krijnse-Locker J. Whole cell cryo-electron tomography reveals distinct disassembly intermediates of vaccinia virus. PLoS ONE. 2007;2:e420. doi: 10.1371/journal.pone.0000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-Dimensional Analysis of a Viral RNA Replication Complex Reveals a Virus-Induced Mini-Organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. First electron tomographic study of a cell infected with a positive-strand RNA virus. The results provide new information on the sites of nascent RNA accumulation and the organization of the RNA replication complex in cells infected with flock house virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baines JD, Hsieh CE, Wills E, Mannella C, Marko M. Electron tomography of nascent herpes simplex virus virions. J Virol. 2007;81:2726–2735. doi: 10.1128/JVI.02571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]