Abstract
The genes for utilization of cellobiose are normally cryptic in both laboratory strains and natural isolates of Escherichia coli. A survey of natural isolates of E. coli reveals that functional genes for cellobiose utilization, while rare, are present. The fraction of E. coli that utilized cellobiose ranged from less than 0.01% in human fecal samples to 7% in fecal samples obtained from horses. Samples obtained from sheep, cows, dogs, and pigs contained 0.1 to 0.5% cellobiose-positive E. coli. Neither the previously identified cel genes nor the bgl genes from E. coli K-12 were expressed during growth on cellobiose by any of the 14 naturally occurring Cel+ isolates that were tested. All of the naturally occurring Cel+ isolates possessed a cel operon, but all were deleted for the major portion of the bgl operon. The functional cel+ genes from these natural isolates differed from the mutationally activated cel+ genes obtained in earlier studies in that (i) the mutationally activated cel+ genes were temperature sensitive, while the functional genes were not, and (ii) transport of cellobiose was inducible in the strains carrying functional cel+ genes, while it was expressed constitutively in strains carrying mutationally activated genes.
Full text
PDF

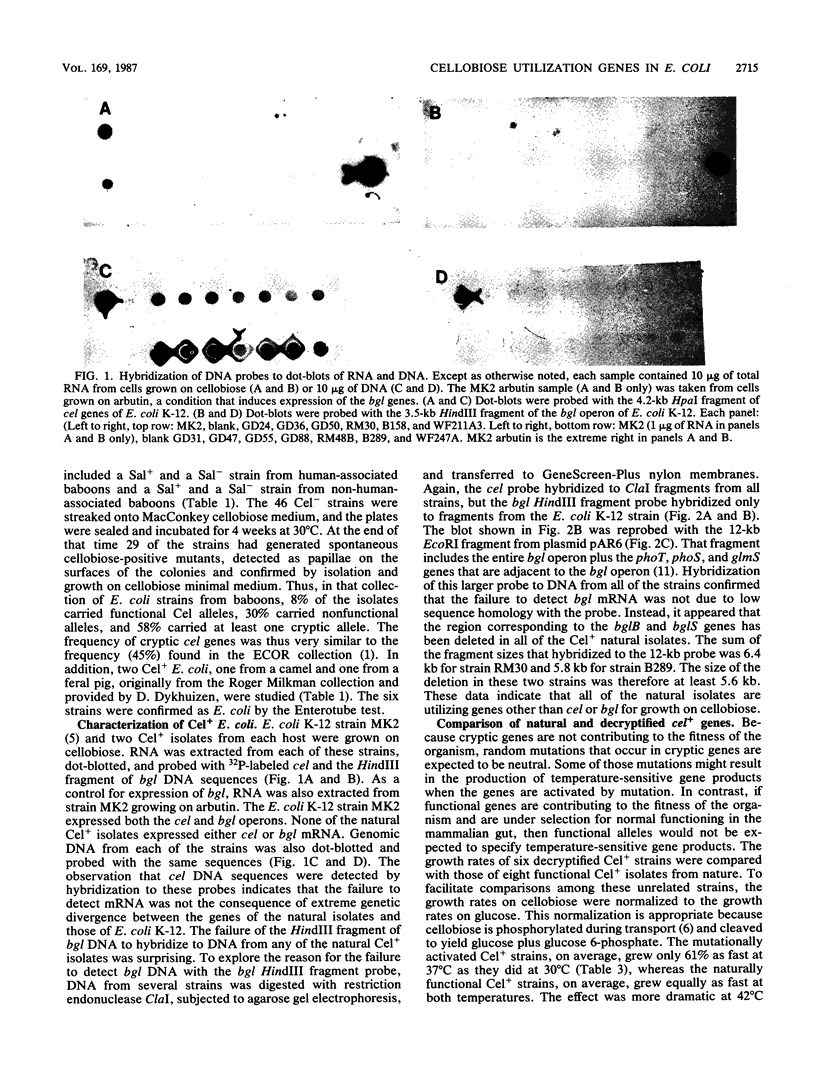


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hall B. G., Betts P. W. Cryptic genes for cellobiose utilization in natural isolates of Escherichia coli. Genetics. 1987 Mar;115(3):431–439. doi: 10.1093/genetics/115.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Betts P. W., Kricker M. Maintenance of the cellobiose utilization genes of Escherichia coli in a cryptic state. Mol Biol Evol. 1986 Sep;3(5):389–402. doi: 10.1093/oxfordjournals.molbev.a040406. [DOI] [PubMed] [Google Scholar]
- Hall B. G., Hartl D. L. Regulation of newly evolved enzymes. I. Selection of a novel lactase regulated by lactose in Escherichia coli. Genetics. 1974 Mar;76(3):391–400. doi: 10.1093/genetics/76.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Yokoyama S., Calhoun D. H. Role of cryptic genes in microbial evolution. Mol Biol Evol. 1983 Dec;1(1):109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- Kricker M., Hall B. G. Biochemical genetics of the cryptic gene system for cellobiose utilization in Escherichia coli K12. Genetics. 1987 Mar;115(3):419–429. doi: 10.1093/genetics/115.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kricker M., Hall B. G. Directed evolution of cellobiose utilization in Escherichia coli K12. Mol Biol Evol. 1984 Feb;1(2):171–182. doi: 10.1093/oxfordjournals.molbev.a040310. [DOI] [PubMed] [Google Scholar]
- Milkman R. Electrophoretic variation in Escherichia coli from natural sources. Science. 1973 Dec 7;182(4116):1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C. E., Williamson V. M. Ty insertions at two loci account for most of the spontaneous antimycin A resistance mutations during growth at 15 degrees C of Saccharomyces cerevisiae strains lacking ADH1. Mol Cell Biol. 1986 Jan;6(1):70–79. doi: 10.1128/mcb.6.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routman E., Miller R. D., Phillips-Conroy J., Hartl D. L. Antibiotic resistance and population structure in Escherichia coli from free-ranging African yellow baboons. Appl Environ Microbiol. 1985 Oct;50(4):749–754. doi: 10.1128/aem.50.4.749-754.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefler S. Inducible system for the utilization of beta-glucosides in Escherichia coli. I. Active transport and utilization of beta-glucosides. J Bacteriol. 1967 Jan;93(1):254–263. doi: 10.1128/jb.93.1.254-263.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefler S., Maas W. K. Inducible system for the utilization of beta-glucosides in Escherichia coli. II. Description of mutant types and genetic analysis. J Bacteriol. 1967 Jan;93(1):264–272. doi: 10.1128/jb.93.1.264-272.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Levin B. R. Genetic diversity and structure in Escherichia coli populations. Science. 1980 Oct 31;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]




