Abstract
A 155-kDa proteinase inhibitor, pacifastin, from plasma of the freshwater crayfish, Pacifastacus leniusculus, was found to be composed of two covalently linked subunits. The two subunits are encoded by two different mRNAs, which were cloned and sequenced. The heavy chain of pacifastin (105 kDa) is related to transferrins, containing three transferrin lobes, two of which seem to be active for iron binding. The light chain of pacifastin (44 kDa) is the inhibitory subunit, and has nine cysteine-rich inhibitory domains that are homologous to each other and to low molecular weight proteinase inhibitors isolated from the grasshopper, Locusta migratoria. The nine light chain domains and the Locusta inhibitors share a characteristic cysteine array (Cys-Xaa9–12-Cys-Xaa2-Cys-Xaa-Cys-Xaa6–8-Cys-Xaa4-Cys) distinct from any described proteinase inhibitor family, suggesting that they constitute a new family of proteinase inhibitors. Pacifastin is the first known protein that has combined properties of a transferrin-like molecule and a proteinase inhibitor.
Serine proteinase inhibitors are widely distributed among living organisms and their major physiological function appears to be to prevent unwanted proteolysis (1–3). A growing number of serine proteinase inhibitors has been described from invertebrates and most of them fall into established inhibitor families such as Kazal-type, Kunitz-type, serpins, Ascaris inhibitors, or α-macroglobulins (4).
Previously we have identified and characterized a 155-kDa proteinase inhibitor from the plasma of the crayfish Pacifastacus leniusculus (5). This protein was found to have an inhibitory activity toward both trypsin and chymotrypsin and is, among the proteinase inhibitors found in crayfish blood, the most efficient inhibitor of the prophenoloxidase activating enzyme of the prophenoloxidase activating system (5, 6), an important immune response cascade in arthropods and many other invertebrates (7). Here we report the purification, characterization, and cDNA cloning of this 155-kDa proteinase inhibitor. This inhibitor is found to be composed of two subunits, a 44-kDa peptide (the light chain) and a 105-kDa peptide (the heavy chain), which are covalently linked. The heavy chain has sequence similarity to transferrins, but in contrast to other transferrins, which have two lobes (8–10), the pacifastin heavy chain has a three-lobe structure. The light chain contains nine homologous cysteine-rich domains with significant similarities to three proteinase inhibitors from L. migratoria (11–14). This is the first known protein that has been found to contain both a transferrin chain and a proteinase inhibitor chain and we have named it pacifastin.
MATERIALS AND METHODS
Animals.
Freshwater crayfish, Pacifastacus leniusculus, were kept in aquaria in aerated tap water at 10°C. Only intermolt males were used.
Purification of Pacifastin.
Pacifastin was purified from crayfish plasma using an established procedure (method 1) described by Aspán et al. (6), and a modified method (method 2) described below. Crayfish blood was collected in an equal volume of ice-cold 10 mM sodium cacodylate, 0.25 M sucrose, 100 mM CaCl2, pH 7.0, and centrifuged (800 × g for 10 min at 10°C) to spin down the blood cells. The supernatant (plasma) was dialyzed overnight against 10 mM Tris⋅HCl buffer (pH 8.0), and centrifuged (5,000 × g for 5 min) to remove precipitates. The supernatant was brought to 0.5 M NaCl and applied to a phenyl-Sepharose column (0.8 cm × 15 cm) equilibrated with 10 mM Tris⋅HCl buffer (pH 8.0), containing 0.5 M NaCl. The column was washed with the same buffer, and the bound proteins were eluted with 10 mM Tris⋅HCl buffer (pH 8.0). Fractions with proteinase inhibitory activity were pooled, dialyzed against 10 mM Tris⋅HCl buffer (pH 8.0), overnight, adjusted to a NaCl concentration of 0.5 M, and fractionated on a second phenyl-Sepharose column under the same conditions. Inhibitor fractions from this column were dialyzed against 10 mM Tris⋅HCl buffer, pH 8.0, and loaded on a DEAE-cellulose column, equilibrated with the same buffer. Bound proteins were eluted by a 0–0.5 M NaCl gradient in 10 mM Tris⋅buffer (pH 8.0). Fractions containing inhibitory activity were pooled and dialyzed against 0.1 M ammonium acetate and lyophilized.
Proteinase Inhibitor Counterstaining.
Proteinase inhibitory activities of intact and proteolytically fragmented pacifastin were visualized after SDS/PAGE using a proteinase inhibitor counterstaining method essentially as described (15, 16). After SDS/PAGE of the samples under reducing and nonreducing conditions, the gel was equilibrated with 0.1 M sodium phosphate buffer (pH 7.8) at 37°C for 15 min, incubated with 40 μg/ml α-chymotrypsin or trypsin (Sigma) for 30 min, and finally developed for 1 h at room temperature in a freshly prepared mixture of 10 ml of 2.5 mg/ml N-acetyl-dl-phenyalanine-β-naphthyl ester in N,N-dimethylformamide and 50 ml 1 mg/ml tetrazotized O-dianisidine in 50 mM sodium phosphate (pH 7.8).
Preparation of a 30-kDa Pacifastin Fragment That Retains Inhibitory Activity.
Two mg of trypsin was immobilized on 0.5 g of CNBr-activated Sepharose 4B (Pharmacia) following the manufacturer’s instructions. These trypsin-Sepharose beads were washed with 0.1 M Tris, pH 8.0, and suspended in 2 ml of the same buffer. Five or ten microliters of this suspension was mixed with 100 μg pacifastin and incubated overnight at 22°C. The mixture was centrifuged at 13,000 × g and the supernatant was collected. Analysis by SDS/PAGE was done for one-twentieth of the samples and the proteinase inhibitor counterstaining method described above. Controls with only pacifastin or only trypsin-Sepharose were incubated and analyzed under identical conditions.
Protein Chemical Procedures.
Five milligrams of pacifastin was CNBr-treated, reduced, and S-carboxymethylated using standard procedures (17). The CNBr fragments of pacifastin were separated on a Superose 12 gel filtration column in 50% HCOOH. Fragments <6 kDa were separated by SDS/PAGE using a 20% Tris⋅tricine gel, and electroblotted onto Problott membranes. After staining with Coomassie brilliant blue (17), individual bands were subjected to N-terminal sequence analysis using an Applied Biosystems model 477A instrument equipped with a model 120A phenylthiohydantoin-derivative analyzer (18). Intact pacifastin was also sequenced after SDS/PAGE and electroblotting.
CNBr fragments smaller than ≈6 kDa were subjected to reverse-phase HPLC on Nucleosil C4 and C18 columns and aqueous trifluoroacetic acid based solvents using acetonitrile or 2-propanol as organic modifiers (17). Pure fragments were spotted onto polybrene-coated glass filters and subjected to sequence analysis.
The 30-kDa pacifastin fragment with proteinase inhibitory activity was resolved on a SDS/polyacrylamide gel under reducing conditions and the peptide band was excised and subjected to in-gel trypsin cleavage (19). The products were fractionated by HPLC and the N-terminal amino acid sequences of three peptides were determined.
Iron Content Determination.
Lyophilized pacifastin was mounted on a carbon specimen holder (Agar Aids, Stansted, U.K.) and coated with a conductive carbon layer. The sample was observed in a Philip 525 scanning electron microscope. X-ray microanalysis was carried out at an accelerating voltage of 20 kV with a Link AN 10000 energy-disperse x-ray microanalysis system. Spectra were acquired for 100 sec. Quantitative analysis was carried out using the link zaf pb program. The results were expressed in mmol of iron per kg dry weight of pacifastin.
Protein Determination.
The concentration of pacifastin was measured with the Bradford method (20). The concentrations of trypsin and chymotrypsin were calculated from absorption values at 280 nm.
Antibodies.
An antiserum was prepared by injecting purified pacifastin intramuscularly into a rabbit. Specific, affinity-purified anti-pacifastin antibodies were concentrated by passing the antiserum over a column of pacifastin-Sepharose (pacifastin crosslinked to CNBr-Sepharose), and eluting the bound antibodies.
Crayfish blood was withdrawn from crayfishes pre-injected with 0.5 ml of a citrate-EDTA anticoagulant solution (21) into syringes containing 1 ml of the same solution. The plasma was separated from blood cells by centrifugation (1,000 × g, 5 min). The pH of the plasma was adjusted to pH 8.0 by adding 2 M Tris⋅HCl (pH 8.0). To verify the specificity of the antibodies, 0.2 μg pacifastin and 20 μg crayfish plasma proteins were run on SDS/PAGE under reducing conditions and electrotransferred to a nitrocellulose membrane. Protein bands recognized by the antibodies were detected by incubating the membrane with the anti-pacifastin antibodies (diluted 1:2,500), followed by peroxidase-conjugated sheep anti-rabbit IgG (United States Biochemical), and staining for peroxidase activity. Control samples were blotted onto nitrocellulose membrane and the protein patterns were visualized by staining with 0.5% amidoblack in water containing 50% methanol and 10% acetic acid.
Immunoscreening of cDNA Clones.
CLIK immunoscreening kit (CLONTECH) and anti-pacifastin antibodies were used to screen a λgt 11 cDNA library from crayfish hepatopancreas (22) in combination with secondary antibodies conjugated to avidin and biotinylated peroxidase. One positive clone containing a 0.8 kb insert was obtained. This insert was amplified with a λgt 11 insert amplification kit, subcloned in a T-vector, prepared according to Marchuk et al. (23), and sequenced using the Sequenase 2.0 kit (United States Biochemical).
PCR Amplification by Using Degenerated Primers.
Two pairs of degenerate, nested primers were designed from the underlined amino acid sequences in a CNBr fragment of pacifastin, MAGYKFNLTQPKNPVRFCVHNEAE. The primers ATG GCN GGN TAY AAR TT (F1) and G GCN GGN TAY AAR TTY AA (F2) in the forward direction were derived from the sequence MAGYKFN, and the primers TCN GCY TCR TTR TGN AC (R1) and TCR TTR TGN ACR CAR AA (R2) in the reverse direction were derived from the sequence FCVHNEAE. F1 and R1 were used to amplify crayfish genomic DNA by PCR (30 cycles of 94°C for 30 sec, 58°C for 30 sec, and 72°C for 45 sec). This PCR (0.1 μl) was reamplified under the same conditions using the F2 and R2 primers. A specific 63-bp product was subcloned into a T-vector and sequenced.
cDNA Screening Using Gene Specific Primers and PCR.
Pacifastin gene-specific primers were selected from the sequence of the 63-bp pacifastin PCR fragment, the 0.8-kb pacifastin cDNA fragment obtained by immunoscreening, and subsequently isolated clones. These primers were used with a pair of nested λgt 11-specific primers (GAC TCC TGG AGC CCG and CCC GTC AGT ATC GGC GG) to obtain additional pacifastin sequences from the hepatopancreas cDNA library by PCRs essentially as described by Gibbons et al. (24). PCRs were performed for 30 cycles at 94°C for 1 min, 58°C for 2 min and 72°C for 2 min. Specifically amplified cDNA fragments were subcloned into T-vectors and sequenced. The reading frames of two independent cDNAs were established by selecting new specific primers from recently obtained pacifastin nucleotide sequences and repeating the PCR, cloning, and sequencing of PCR products, as described above. To reduce the risk of sequence errors due to the PCR amplification procedure, we determined the sequence of at least two independent PCR products. In case of sequence differences, a third PCR was performed and the product sequenced. A majority decision was made—i.e., the nucleotide present in two of the three products was used.
Northern Analysis.
Total RNAs were isolated from crayfish hepatopancreas and hemocytes using the acid-guanidine method (25). Polyadenylylated RNAs were prepared from total RNAs using mRNA purification kit (Pharmacia) according to the manufacturer’s instructions. Polyadenylylated RNA (3 μg) were electrophoretically separated on 1% agarose gels containing formaldehyde and blotted onto Hybond N membranes (Amersham) using standard techniques. cDNA probes were radioactively labeled by using the Megaprime labeling kit (Amersham). Hybridization and washings were carried out at 65°C according to instructions of the manufacturer and the nylon membrane was exposed to x-ray film for 5 days.
RESULTS
Purification, N-Terminal Sequences, and Inhibitory Activity of Pacifastin.
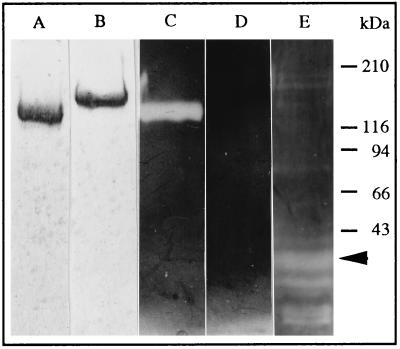
By using method 1, described in Aspán et al. (6) or method 2 (see Material and Methods), we purified a plasma proteinase inhibitor that was present in similar amounts, had the same molecular mass, inhibitory profile, and electrophoretic behavior (Fig. 1, lanes A and B) and it was recognized by the anti-pacifastin antibodies. We therefore conclude that the same protein, pacifastin, was purified using these two methods. The protein prepared according to method 1, had a major N-terminal sequence ASLNLPGRLXVAQ. In cycles 4, 5, and 7, low yields of E, Q, and D, respectively, were also observed. In the protein prepared according to method 2, a major N-terminal sequence ASLEQPD was present. This indicated the presence of two distinct types of subunits (see further below), which appear to become blocked to different extents during the two purification methods. However, only one single band of 155 kDa under reducing conditions (Fig. 1, lane B) and 140 kDa under nonreducing conditions (Fig. 1, lane A) was seen on SDS/PAGE.
Figure 1.
Samples of pacifastin (5 μg) were subjected to SDS/PAGE under nonreducing (lanes A, C, and E) or reducing (lanes B and D) conditions. The gels were Coomassie stained (lanes A and B) or counterstained for chymotrypsin inhibitory activity (lanes C, D, and E). The sample in lane E was preincubated with immobilized trypsin, and a 30-kDa fragment with strong chymotrypsin inhibitory activity is indicated by an arrow.
Pacifastin was previously shown to have inhibitory activities toward trypsin and chymotrypsin (5). These inhibitory activities were found to be retained after treatment with 1% SDS for 5 min at 100°C in the absence of a reducing agent, followed by SDS/PAGE (Fig. 1, lanes C and E). But treatment under the same conditions in the presence of 100 mM dithiothreitol rendered pacifastin completely inactive (Fig. 1, lane D). These results show that the integrity of disulfide bridges is essential for the inhibitory activity of pacifastin. After interaction with trypsin, pacifastin degraded into smaller molecules. Some of the fragments, ranging from 10 to 70 kDa, were shown to retain inhibitory activity against chymotrypsin (Fig. 1, lane E). A peptide of 30 kDa was one of the most abundant degradation products with a strong inhibitory activity.
Cloning and Sequence Analysis of the Light and Heavy Chains of Pacifastin.
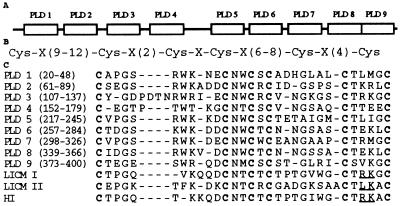
Sequence databases were searched using the blast programs (26). From a crayfish hepatopancreas cDNA library we obtained a clone of 0.8 kb by immunoscreening with anti-pacifastin antibodies. PCR was employed to establish a cDNA of 1,456 bp. This cDNA has an ORF encoding a protein of 420 amino acids and was named the pacifastin light chain. The N-terminal sequence of pacifastin prepared with method 2 is found in the deduced amino acid sequence, showing that it contains a putative signal peptide of 15 amino acids, and a mature protein consisting of 405 amino acids with a calculated molecular mass of 44 kDa (Fig. 2A). Three internal peptide sequences, obtained from the 30-kDa fragment exhibiting inhibitory activity (Fig. 1, lane E), was present in the deduced sequence. The protein has nine cysteine-rich stretches that are homologous to each other, indicating the presence of nine domains (Fig. 3A). Pairwise comparisons showed that the identities between the nine pacifastin light chain domains (PLDs) fall in a range from 76% to 34%. Each PLD has six cysteines arranged in a characteristic spacing pattern (Cys-Xaa9–12-Cys-Xaa2-Cys-Xaa-Cys-Xaa6–8-Cys-Xaa4-Cys) (Fig. 3B). The PLDs share significant sequence similarities with the low molecular weight proteinase inhibitors LICM I, LICM II (11, 12), and HI (14) isolated from the grasshopper L. migratoria (Fig. 3C).
Figure 2.
cDNA sequence and deduced amino acid sequence of crayfish pacifastin light chain (A) and heavy chain (B). Nucleotides and amino acids are numbered to the right. The putative signal peptide sequences are in italic. Underlined sequences were determined by peptide sequencing. Putative N-glycosylation sites are marked in bold.
Figure 3.
Domain structure of the pacifastin light chain. (A) Schematic diagram showing the positions of the nine PLDs. (B) The common cysteine spacing pattern of the nine PLDs, and LICM I, LICM II (11), HI (14). (C) Alignment of the nine PLDs, LICM I, LICM II, and HI. Conserved cysteines are in boldface. Underlined residues indicate putative reactive site, P1-P′1 of the three locust inhibitors (14). Gaps (−) are introduced to optimize the alignment.
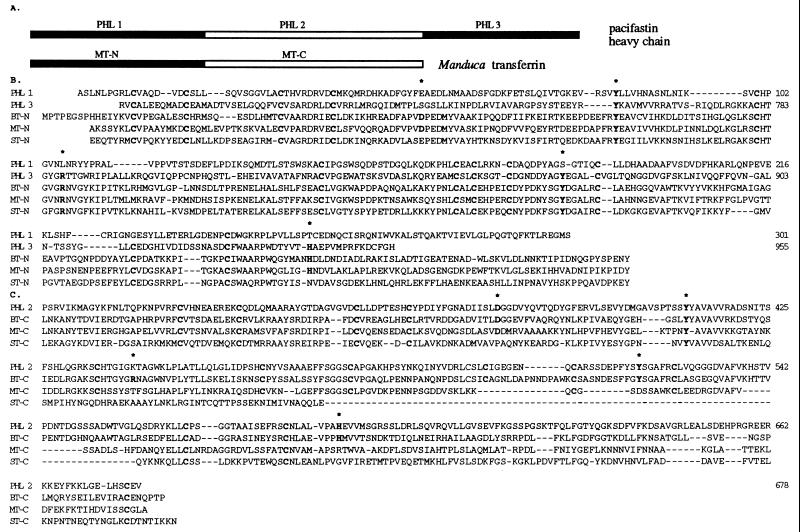
A large number of sequences from CNBr-peptides of pacifastin were not part of the pacifastin light chain. From the sequence of one of these peptides (see Materials and Methods), a 63-bp genomic DNA fragment was obtained by PCR with two pairs of nested degenerate primers. This fragment was found to contain the coding sequence of the pacifastin peptide (AGYKFNLTQPKNPVRFCVHNE). Gene-specific primers were synthesized according to this 63-bp fragment and the PCR products subsequently obtained from the hepatopancreas cDNA library by PCR. We established a cDNA of 3,029 bp, with an ORF encoding a protein of 977 amino acids (Fig. 2B). The protein was confirmed to be the second subunit of pacifastin by the sequence of CNBr fragments of pacifastin and the N-terminal sequence of pacifastin prepared with method 1. The deduced amino acid sequence consists of a putative signal sequence of 22 amino acids and a mature protein of 955 amino acids with an estimated molecular mass of 105 kDa, which was designated the heavy chain of pacifastin. The protein sequence was found to consist of three pacifastin heavy chain lobes (PHLs), with sequence similarity to each other (Fig. 4 A and B), and to transferrins from different animals (8–10). PHL 1 and PHL 3 have highest similarity to the N-terminal lobe of transferrins and PHL 2 is most similar to the transferrin C-terminal lobe (8–10) (Fig. 4 A and B). Crystallographic studies of human lactoferrin and chicken transferrin have indicated that five amino acid residues are necessary for the iron binding in transferrins (27, 28). In PHL 2 and PHL 3, four of these residues were conserved (D379, Y411, Y520, H589 in PHL 2; Y758, R787, Y870, H941 in PHL 3), while in PHL 1, only one (Y85) of these residues was conserved (Fig. 4B). This suggests that PHL 2 and PHL 3 may be active for metal binding.
Figure 4.
Structure of the pacifastin heavy chain. (A) Schematic diagram showing the three PHLs in comparison with Manduca transferrin (8). Filled boxes represent sequence similarity to the N-terminal lobe of transferrins and open boxes represent similarity to C-terminal lobe of transferrins. (B) Alignment of PHL 1 and PHL 3 with N-terminal lobe of transferrins from B. discoidalis, BT-N (9), M. sexta, MT-N (8), and S. peregrina, ST-N (10). (C) Alignment of PHL 2 with C-terminal lobe of transferrins from B. discoidalis (BT-C), M. sexta (MT-C), and S. peregrina (ST-C). Conserved residues that are essential for iron-binding are in bold and indicated with an ∗. Conserved cysteines are in bold. Gaps (−) are introduced to optimize the alignment.
Determination of the Iron Content of Pacifastin.
The iron content of purified pacifastin was determined to be 8.8 mmol/kg protein, which represents a molar ratio of iron to pacifastin of 1.35 to 1. This indicates at least one active iron binding site in pacifastin. Correlating this result with the sequence analysis, it is quite likely that PHL 2 and PHL 3 are both active for iron binding, but their binding sites are not saturated in the purified pacifastin.
Nature of the Crosslink Between the Heavy and Light Chains.
Boiling pacifastin with reducing agents does not dissociate the subunits (Fig. 1, lane B), indicating a covalent link other than disulfide bonds. The link appears not to contain carbohydrates, as treatments with trifluoromethane sulfonic acid, hyaluronidase, or dilute NaOH had no effect. The nature of the bond could not be determined, but from the sequencing of CNBr-fragments we obtained indications of its location. After gel filtration of pacifastin CNBr-fragments on a Superose 12 column, followed by SDS/PAGE and blotting to polyvinylidene difluoride membranes, bands were cut out from the membrane and sequenced. One major band, ≈32 kDa, was found to contain two amino acid sequences present in approximately equal amounts (Fig. 5). The two sequences corresponded to the N-terminals of one CNBr-fragment from the light chain (residues 325–385, ≈7 kDa) and one CNBr-fragment from the heavy chain (residues 403–593, ≈20 kDa). Their appearance together with a size of ≈32 kDa is best explained by a covalent link between these two fragments. Both CNBr-fragments contain putative N-glycosylation sites, which can explain the slightly larger size than expected in SDS/PAGE.
Figure 5.
N-terminal sequence of a 32-kDa CNBr-fragment of pacifastin. Two sequences were present in approximately equal amounts (A). The sequences were identified as the N-terminals of one light chain CNBr-fragment (B, residues 325–385) and one heavy chain CNBr-fragment (C, residues 403–593), indicating a covalent link between the two CNBr-fragments.
Expression of Pacifastin.
The expression of the two chains of pacifastin in hepatopancreas and hemocytes was studied by Northern analysis (data not shown). A mRNA of 5.0 kb was detected with the heavy chain specific probe. The light chain probe detected two transcripts, with sizes of 2.2 and 4.5 kb. All transcripts could be detected in RNA preparations from hepatopancreas as well as hemocytes, even though the signals obtained with the light chain probe were very weak in hemocytes. This indicates that hepatopancreas and hemocytes can synthesize both the heavy chain and light chain of pacifastin. However, it is unclear whether the two transcripts detected by the light chain specific probe are transcription products of the same gene or if one of them represents another highly similar gene product .
DISCUSSION
In this paper we report the cloning and characterization of a novel 155-kDa proteinase inhibitor that we have named pacifastin. This protein is a heterodimeric protein, consisting of one proteinase inhibitory light chain, and one heavy chain related to transferrins. Peptide sequences obtained from the 155-kDa band covered a total of 8% of the light chain (34 of 405 amino acids), and 23% of the heavy chain (217 out of 955 amino acids), confirming the identity of the two cloned sequences. The subunits are covalently linked, but not by disulfide bonds (Fig. 1, lane B). The covalent association is not due to any artifact during handling. We detected no free forms of pacifastin light or heavy chains in crayfish plasma using anti-pacifastin antibodies, and no low molecular mass proteinase inhibitory activity can be detected in SDS/PAGE with the proteinase counterstaining method. Injecting a citrate-EDTA anticoagulant into the crayfish prior to bleeding, to prevent the degranulation and lysis of crayfish blood cells, and the subsequent transglutaminase-mediated clotting (29), did not have any effect on the molecular properties of pacifastin, showing that the transglutaminase activity associated with clotting is not responsible for linking the pacifastin subunits together. Treatments with trifluoromethane sulfonic acid, hyaluronidase, and dilute NaOH (16) can break the carbohydrate chain between the subunits of the inter-α-proteinase inhibitors, but these reagents were unable to affect the linkage between the two subunits of pacifastin. It appears therefore that the covalent linkage in pacifastin is different from that in the inter-α-trypsin inhibitors.
The only known family of heteropolymeric proteinase inhibitors are the mammalian inter-α-trypsin inhibitors (30, 31), which are composed of Kunitz-type proteinase inhibitor subunits (bikunin) and one or two heavy subunits, linked together by a carbohydrate chain. The heavy subunits have recently been implicated to be related to proteins of the multicopper oxidase family (32). The bikunin was also found to be present in free form in human serum and urine (33).
The heavy chain of pacifastin has significant similarity to transferrins from other animals (Fig. 4). Transferrins consist of two distinct lobes, which correspond to the N-terminal half and the C-terminal half of the protein. The term lobe is used instead of domain, as the lobe may have domains within it. Each lobe folds independently to form an iron-binding site (27, 28). It is widely accepted that two-lobe transferrins have arisen from a single-lobe transferrin ancestor through gene duplication (34). This two-lobe structure applies to all members of the transferrin family (35) with the exception of hemiferrin, in which only a single iron-binding lobe is present (36). The pacifastin heavy chain, however, is a novel member of the transferrin family, which has a previously unseen three-lobe structure. Two of the lobes, PHL 1 and 3, are similar to the N-terminal lobe of transferrins, and PHL 2 is similar to the C-terminal lobe. Four of five residues necessary for iron binding were found to be conserved in PHL 2 and 3 (Fig. 4B) and it is quite likely that both these lobes are capable of binding iron. By measuring the iron content, it was shown that pacifastin has at least one, probably two, active iron binding site(s). Thus pacifastin is not only structurally but also functionally related to transferrins. The size of pacifastin is reminiscent of a transferrin that has been purified from another crustacean, the crab Cancer magister (37). This crab protein has a molecular mass of 150 kDa and contains two iron-binding sites, but it was not analyzed for proteinase inhibitor activity. It is likely that this crab transferrin is a homologue of crayfish pacifastin, although sequence data of this protein to confirm this proposal is still lacking.
In the first lobe of the pacifastin heavy chain (PHL 1, Fig. 4B), only one of the five amino acid residues important for iron binding was conserved, indicating that this lobe may not be able to form an iron-binding site. Two of the three cloned insect transferrins also contain such an inactive lobe (8–10). Saxiphilin (38), another transferrin-like protein purified and cloned from bullfrog, Rana catesbeiana, was found to consist of two lobes that have both lost their iron-binding activity and have acquired affinity for saxitoxin, a neurotoxin produced by various dinofagellates and cyanobacteria (39). It is not known if the inactive lobes of invertebrate transferrins have evolved to have other functions than iron-binding.
The light chain of pacifastin was found to contain nine homologous domains (PLDs) that also are homologous to three low molecular weight proteinase inhibitors (LICM I, LICM II, and HI) that were isolated from the grasshopper L. migratoria (11, 12, 14) (Fig. 3 A and C). The PLDs also show sequence similarity to proteins with cysteine-rich stretches, like von Willebrand factor, a mammalian blood protein involved in coagulation (40), proteins involved in development, like the Jagged protein (41), and a cloned sequence of unknown function from C. elegans (42). The sequence similarity does not depend solely on the pattern of the cysteine residues, but none of these proteins are known to be proteinase inhibitors. It is therefore difficult to say if the similarities are coincidental or not. The two proteinase inhibitors LICM I and LICM II are derived from a two-domain precursor encoded by a single mRNA (13), indicating a posttranslational processing into two single domain proteinase inhibitors. The pacifastin light chain does not seem to be processed into single domain peptides. Each PLD and LICM I/LICM II/HI contains six cysteine residues with the same spacing pattern, Cys-Xaa9–12-Cys-Xaa2-Cys-Xaa-Cys-Xaa6–8-Cys-Xaa4-Cys (Fig. 3B). This pattern is distinct from any other recognized proteinase inhibitor family (1–3), suggesting that these inhibitors constitute a new family of proteinase inhibitors. Kellenberger et al. (14) places the reactive site P1-P′1 of the three Locusta inhibitors between the two last cysteines, close to the C-terminal end. Variations in these P1-P′1 residues were shown to effect the inhibitory specificity. The corresponding residues in the nine aligned crayfish PLDs show variations (Fig. 3C), indicating that the different domains could be specific inhibitors for different proteinases. The inhibitory activity of pacifastin toward chymotrypsin, trypsin, and also elastase (5), could therefore reside in different domains. This family of proteinase inhibitors is so far only found in arthropods. In addition, crayfish pacifastin is a novel type of proteinase inhibitor in which a transferrin subunit is linked to a proteinase inhibitory subunit.
Acknowledgments
We thank Dr. Lage Cerenius for discussions and help, Zhengzhu Jin for performing iron content determination, Ragnar Ajaxon, and Lene Kristensen for technical assistance. This work has been supported by grants to K.S. from the Swedish Natural Science Research Council and the Swedish Forestry and Agricultural Research Council, and to L.S.-J. from the Danish Biomembrane Research Center.
ABBREVIATIONS
- PHL
pacifastin heavy chain lobe
- PLD
pacifastin light chain domain
Footnotes
References
- 1.Laskowski M, Jr, Kato I. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 2.Travis J, Salvesen G S. Annu Rev Biochem. 1983;52:655–709. doi: 10.1146/annurev.bi.52.070183.003255. [DOI] [PubMed] [Google Scholar]
- 3.Bode W, Huber R. Eur J Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- 4.Kanost M R, Jiang H. In: New Directions in Invertebrate Immunity. Söderhäll K, Iwanaga S, Vasta G R, editors. Fair Haven, NJ: SOS; 1996. pp. 155–173. [Google Scholar]
- 5.Hergenhahn H-G, Aspán A, Söderhäll K. Biochem J. 1987;248:223–228. doi: 10.1042/bj2480223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aspán A, Hall M, Söderhäll K. Insect Biochem. 1990;20:458–492. [Google Scholar]
- 7.Söderhäll K, Cerenius L, Johansson M W. In: New Directions in Invertebrate Immunity. Söderhäll K, Iwanaga S, Vasta G R, editors. Fair Haven, NJ: SOS; 1996. pp. 155–173. [Google Scholar]
- 8.Bartfeld N S, Law J H. J Biol Chem. 1990;265:21684–21691. [PubMed] [Google Scholar]
- 9.Kurama T, Kurata S, Natori S. Eur J Biochem. 1995;228:229–235. [PubMed] [Google Scholar]
- 10.Jamroz R C, Gasdaska J R, Bradfield J Y, Law J H. Proc Natl Acad Sci USA. 1993;90:1320–1324. doi: 10.1073/pnas.90.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boigegrain R-A, Mattras H, Brehélin M, Paroutaud P, Coletti-Previero M-A. Biochem Biophys Res Commun. 1992;189:790–793. doi: 10.1016/0006-291x(92)92271-x. [DOI] [PubMed] [Google Scholar]
- 12.Nakakura N, Hietter H, van Dorsselaer A, Luu B. Eur J Biochem. 1992;204:147–153. doi: 10.1111/j.1432-1033.1992.tb16617.x. [DOI] [PubMed] [Google Scholar]
- 13.Kromer E, Nakakura N, Lagueux M. Insect Biochem Mol Biol. 1994;24:329–331. doi: 10.1016/0965-1748(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 14.Kellenberger C, Boudier C, Bermudez I, Bieth J, Luu B, Hietter H. J Biol Chem. 1995;270:25514–25519. doi: 10.1074/jbc.270.43.25514. [DOI] [PubMed] [Google Scholar]
- 15.Uriel J, Berges J. Nature (London) 1968;218:578–580. doi: 10.1038/218578b0. [DOI] [PubMed] [Google Scholar]
- 16.Enghild J J, Thøgersen I B, Pizzo S V, Salvesen G. J Biol Chem. 1989;264:15975–15981. [PubMed] [Google Scholar]
- 17.Matsudaira P T. A Practical Guide to Protein and Peptide Purification for Microsequencing. Orlando, FL: Academic; 1989. [Google Scholar]
- 18.Sottrup-Jensen L. Anal Biochem. 1995;225:187–188. doi: 10.1006/abio.1995.1137. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld J, Capdevielle J, Guillemot J C, Ferrara P. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Smith V J, Söderhäll K. Cell Tissue Res. 1983;233:295–303. doi: 10.1007/BF00238297. [DOI] [PubMed] [Google Scholar]
- 22.Cerenius L, Liang Z, Duvic B, Keyser P, Hellman U, Palva E P, Iwanaga S, Söderhäll K. J Biol Chem. 1994;269:29462–29467. [PubMed] [Google Scholar]
- 23.Marchuk D, Drumm M, Saulino A, Collins F S. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons I R, Asai D J, Ching N S, Dolecki G J, Mocz G, Philipson C A, Ren H, Tan W-J, Gibbons B H. Proc Natl Acad Sci USA. 1991;88:8563–8567. doi: 10.1073/pnas.88.19.8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.&erson B F, Baker H M, Norris G E, Rice D W, Baker E N. J Mol Biol. 1989;209:711–734. doi: 10.1016/0022-2836(89)90602-5. [DOI] [PubMed] [Google Scholar]
- 28.Bailey S, Evans R W, Garratt R C, Gorinsky B, Hasnain S, Horsburg C, Jhoti H, Lindey P F, Mydin A, Sarra R, Watson J L. Biochemistry. 1988;27:5804–5812. doi: 10.1021/bi00415a061. [DOI] [PubMed] [Google Scholar]
- 29.Kopacek P, Hall M, Söderhäll K. Eur J Biochem. 1993;213:591–597. doi: 10.1111/j.1432-1033.1993.tb17798.x. [DOI] [PubMed] [Google Scholar]
- 30.Bourguignon J, Diarra-Mehrpour M, Thiberville L, Bost F, Sesboüé R, Martin J P. Eur J Biochem. 1993;212:771–776. doi: 10.1111/j.1432-1033.1993.tb17717.x. [DOI] [PubMed] [Google Scholar]
- 31.Diarra-Mehrpour M, Bourguignon J, Bost F, Sesboüé R, Muschio F, Sarafan N, Martin J-P. Biochim Biophys Acta. 1992;1132:114–118. doi: 10.1016/0167-4781(92)90065-8. [DOI] [PubMed] [Google Scholar]
- 32.Chan P, Risler J L, Salier J P. Biochem J. 1995;306:505–512. doi: 10.1042/bj3060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salier J P. Trends Biochem Sci. 1990;15:435–439. doi: 10.1016/0968-0004(90)90282-g. [DOI] [PubMed] [Google Scholar]
- 34.Williams J. Trends Biochem Sci. 1982;7:394–397. [Google Scholar]
- 35.Baldwin G S. Comp Biochem Physiol. 1993;106B:203–218. doi: 10.1016/0305-0491(93)90028-4. [DOI] [PubMed] [Google Scholar]
- 36.Stallard B J, Collard M W, Griswold M D. Mol Cell Biol. 1991;11:1448–1453. doi: 10.1128/mcb.11.3.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huebers H A, Huebers E, Finch C A, Martin A W. J Comp Physiol. 1982;148:101–109. [Google Scholar]
- 38.Morabito M A, Moczydlowski E. Proc Natl Acad Sci USA. 1994;91:2478–2482. doi: 10.1073/pnas.91.7.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall S, Reichardt P B. Marine Toxins: Origin, Structure and Molecular Pharmacology. Washington, DC: Am. Chem. Soc.; 1990. [Google Scholar]
- 40.Sadler J E. J Biol Chem. 1991;266:22777–22780. [PubMed] [Google Scholar]
- 41.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 42.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]