Abstract
We have developed a new biochemical method to isolate a homogeneous population of RNA polymerase II (RNA pol II) elongation complexes arrested at a DNA damage site. The method involves triple-helix formation at a predetermined site in DNA template with a third strand labeled with psoralen at its 5′-end and a biotin at the 3′-end. After triplex formation and near-ultraviolet irradiation (360 nm), DNA templates modified with psoralen were immobilized on streptavidin-coated magnetic beads and used for in vitro transcription reactions with HeLa nuclear extracts. Separation of magnetic beads from solution results in isolation of arrested elongation complexes on the immobilized DNA templates. We have applied the method to arrest RNA pol II elongation complexes on a DNA template containing HIV-1 promoter. Our results indicate that psoralen crosslink in the template strand efficiently arrests elongation complexes, and psoralen monoadducts terminate transcription. Our results also demonstrate that a triple-helical structure stabilized by an intercalator, acridine, attached to the third strand of the helix inhibits transcription by a termination pathway. Isolation of stable RNA pol II elongation complexes arrested at DNA damage sites is a remarkable finding. This result demonstrates that arrested elongation complexes are impervious to DNA damage repair machinery and other regulatory proteins present in HeLa nuclear extracts. The method of delivering site-specific psoralen damage by a triplex structure and isolation of arrested RNA pol II elongation complexes should be generalizable to any promoter and DNA template sequences. This strategy provides a new approach to study the mechanism of transcription elongation and transcription-coupled DNA damage repair.
Keywords: psoralen photocrosslinking, triple helix, DNA repair
Elongation stage of eukaryotic mRNA synthesis is an important target for regulation of gene expression. Expression of many cellular genes and the growth of many viruses, including HIV type 1 (HIV-1) and adenovirus involve regulation of transcription elongation and termination (1, 2). Recent studies suggest that general and regulatory elongation factors are found in eukaryotes (3). During elongation, RNA polymerase II (RNA pol II) can pause, get arrested, pass through terminator sequences, or terminate transcription. To understand elongation and termination stages of RNA pol II transcription, structural characterization of elongation complexes halted at specific sites during transcription would be very useful (4) as has been done for ternary complexes of T7 and Escherichia coli RNA polymerases halted during transcription (5–10).
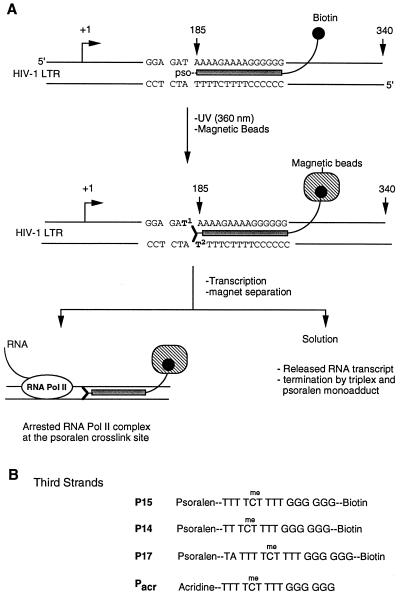
Psoralen adducts site-specifically placed in DNA templates have been used to arrest T7 and E. coli RNA polymerase elongation complexes for structural characterization (5–8). We reasoned that, as with T7 and E. coli RNA polymerases, RNA pol II elongation complexes can be arrested by introducing a psoralen adduct at a specific site in DNA templates. Triple-helical DNA structures were used to deliver site-specific psoralen adducts in the DNA template (11). As a model system, we introduced psoralen adducts in a DNA template containing HIV-1 promoter. The experimental strategy for site-specific damage in the DNA template containing HIV-1 promoter and isolation of the arrested RNA pol II elongation complexes is outlined in Fig. 1. Our method involves the following six steps: (i) insertion of a target sequence for triple-helix formation at a predetermined position in the DNA template; (ii) synthesis of a third strand for triplex formation containing a psoralen at its 5′-end and a biotin at the 3′-end; (iii) triplex formation between the DNA template and the third strand followed by near-ultraviolet irradiation (360 nm); (iv) immobilization of the crosslinked DNA template on streptavidin-conjugated magnetic beads and in vitro transcription; (v) separation of magnetic beads from solution; and (vi) analysis of the RNA transcripts and arrested RNA pol II complexes by gel electrophoresis and immunoblotting.
Figure 1.
(A) Experimental design to isolate a homogeneous population of RNA pol II elongation complexes and to study the effects of DNA damage on transcription elongation. A target sequence for triple-helical DNA is inserted into the DNA template. A psoralen and biotin containing oligonucleotide is used to form triplex DNA, and UV irradiation covalently crosslinks psoralen to the template. Psoralen-crosslinked template is immobilized on streptavidin-conjugated magnetic beads, and noncrosslinked DNA is washed away with buffer. Cell-free transcription reactions are performed, and beads are separated from solution by a magnet. The protein composition of the arrested complexes is analyzed by immunoblotting. Different T residues in the target sequence are numbered. Arrows and numbers at +1, 185, and 340 indicate transcriptional start, triplex, and runoff sites, respectively. (B) Structures of third-strand probes used to form triple-helices with the DNA template outlined above. All of these oligonucleotides were modified with psoralen or acridine at their 5′-end. Three probes contained biotin at their 3′-end. C residues in the sequences were substituted with 5-methyl-C to stabilize triplex structures.
Four DNA strands conjugated with psoralen and acridine were synthesized to form triple-helical structures with the DNA template and their effect on transcription has been studied. Our results revealed that psoralen crosslinks efficiently arrested RNA pol II elongation complexes, whereas psoralen monoadducts on the template strand and stable triple-helical DNA terminated transcription. Transcriptional arrest and termination sites were determined by S1 nuclease mapping of the 3′-end of RNA transcripts. These experiments provide a new approach to isolate a homogeneous population of RNA pol II elongation complexes required to investigate the polymerase–DNA interactions during transcription and to study the mechanism of transcription inhibition by triplex DNA structures.
MATERIALS AND METHODS
Template DNAs.
The test plasmid (pWT-1) used in this study was derived from the p10SLT plasmid that contains HIV-1 5′ long terminal repeat (12). Plasmid pWT-1 was constructed by inserting a synthesized DNA fragment containing a triplex target sequence (5′-AAA AGA AAA GGG GGG-3′) between HindIII and NarI sites of plasmid p10SLT. Templates were linearized by cleavage with AccI (+340). To avoid multiple additions of psoralen, a GC-rich sequence was introduced upstream from the triplex site.
Triplex Formation and Crosslinking.
Linearized pWT-1 DNA (0.1 μM final concentration) was incubated with excess psoralen-oligonucleotide probe (×500) in a buffer containing 10 mM Tris⋅HCl at pH 6.5, 50 mM NaCl, 10 mM MgCl2, and 0.5 mM spermine for 30 min at 37°C, then cooled down slowly to 0°C (≈30 min). The mixture was irradiated with UV (360 nm) on ice in a Photochemical Reactor for 15 min. For magnetic-bead binding, template DNA was separated from unreacted psoralen-oligonucleotide probe on 1% agarose gels.
Cell-Free Transcription Experiments.
HeLa cell nuclear extracts were prepared according to published procedures (13, 14). Template DNAs were linearized with AccI. Transcription reaction mixtures (25 μl) contained 0.5 μg of template DNA, 12 μl of HeLa cell nuclear extract, 50 mM KCl, 6 mM MgCl2, 10 mM Hepes at pH 7.9, 1 mM DTT, 10 mM phosphocreatine, 80 μM CTP, 80 μM ATP, 80 μM UTP, and 10 μCi of [α-32P]GTP (25 Ci/mmol, ICN; final concentration, 20 μM). Psoralen- and acridine-labeled DNA strands were added to final concentrations as indicated in Figs. 2, 3, 4. After incubation for 50 min at 30°C, 175 μl of stop solution (0.3 M Tris⋅HCl, pH 7.4/0.3 M sodium acetate/0.5% SDS/2 mM EDTA) was added. The mixture was extracted with 200 μl of phenol/chloroform/isoamyl alcohol (50:48:2) and then with chloroform (200 μl). RNA transcripts were precipitated with ethanol and analyzed on 6% polyacrylamide/7 M urea gels.
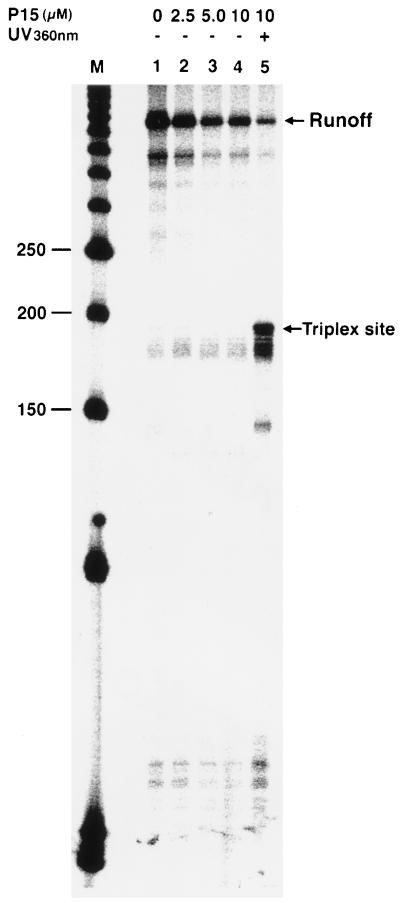
Figure 2.
Specific inhibition of transcription elongation by psoralen adducts at the triplex site. Lane 1 (control), transcription without third strand. Lanes 2–4, increasing concentration of third-strand P15 without UV crosslinking. Lane 5, addition of 10 μM P15 and photocrosslinking reaction followed by transcription. Lane M is a 50-bp DNA marker, and nucleotide lengths (150 to 250) around the triplex site are shown.
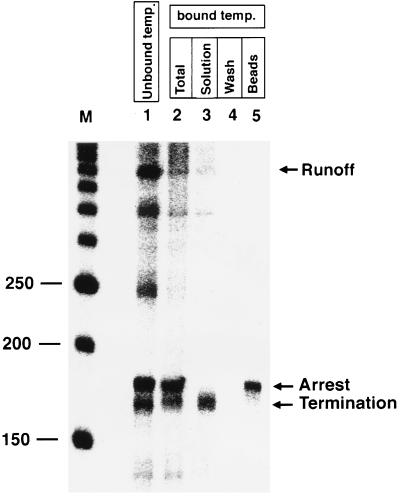
Figure 3.
Isolation of RNA pol II elongation complexes arrested at the DNA damage site. Lane 1, P15 (10 μM) was used to form triplex, UV irradiated, and used for transcription. Lane 2, triplex was formed with P15 (10 μM), UV irradiated, and crosslinked DNA template was immobilized on magnetic beads and used for transcription. Lanes 3–5, transcription reactions were performed as in lane 2 and separated by magnet: solution phase (lane 3); wash with buffer D (lane 4); and beads treated with stop solution to release the bound RNA pol II complex (lane 5). Lane M is a 50-bp DNA marker, and nucleotide lengths (150 to 250) around the triplex site are shown. RNA transcripts in the solution phase and bound to the beads are labeled as termination and arrest, respectively.
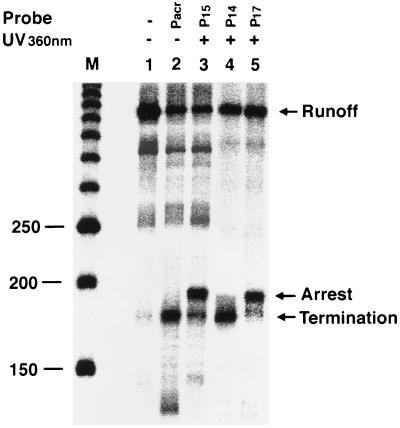
Figure 4.
Arrest and termination of RNA pol II elongation complexes by various oligonucleotide probes. Third-strand probes (10 μM) were added to the transcription reaction and UV irradiated as indicated (not isolated on magnetic beads). Lane M is a 50-bp DNA marker, and nucleotide lengths (150 to 250) around the triplex site are shown. Transcripts arising from transcription termination and arrest are indicated. Full-length transcripts produced from unmodified templates are also shown.
For DNA templates immobilized on magnetic beads, transcription was performed for 50 min, and then beads were separated from nuclear extract by magnetic particle concentrator (Dynal, Great Neck, NY). The recovered beads containing arrested elongation complexes were washed three times with 50 μl of buffer D (20 mM Hepes, pH 7.9/0.5 mM DTT/100 mM KCl/20% glycerol/0.2 mM EDTA). To release bound RNA transcripts and proteins from the beads, 200 μl of stop solution was added to the beads, and the mixture was incubated for 10 min at 37°C followed by magnetic separation. The solution phase contained released RNA transcripts and proteins. RNA transcripts were analyzed as described above.
RESULTS
Transcription Elongation by RNA Pol II Is Inhibited by a Site-Specific Psoralen Adduct in DNA Template.
Triple-helix-forming oligonucleotides were modified with psoralen and used to induce site-specific damage in HIV-1 promoter sequence. The effect of a psoralen-containing oligonucleotide probe, P15 (Fig. 1B), was investigated by in vitro transcription experiments, and transcript products were analyzed by denaturing gel electrophoresis. Results of these experiments are shown in Fig. 2. Covalent crosslinking of P15 to the target site by irradiation caused transcription inhibition at the triplex site and produced short-length transcripts as the major product and a reduced amount of runoff transcripts (Fig. 2, lane 5). Without the psoralen photocrosslinking to the target site, no transcription inhibition was observed at the triplex site (Fig. 2, lanes 2–5). However, at higher concentrations of probe P15, the overall level of transcription was lower and a decrease in the runoff transcripts was observed. These results indicate that a psoralen adduct can block an elongation RNA pol II complex. Our results further show that triple-helical structures formed by P15 are not stable during in vitro transcription conditions and cannot stop RNA pol II at specific sites.
Psoralen Adducts Can Arrest and Terminate RNA Pol II Elongation Complexes.
To address the question of whether the transcription inhibition by psoralen probe P15 crosslinked to the template is the result of RNA pol II elongation complexes arrest at the damage site or the result of transcript release and termination, we developed a new strategy to isolate arrested RNA pol II complexes (Fig. 1). We used third-strand P15 to form a triplex at the target site, UV irradiated at 360 nm, purified psoralen-modified and unmodified template DNA from excess P15 by gel, treated this mixture of DNA templates with streptavidin-coated beads, extensively washed beads with buffer to remove unmodified DNA template, and used this immobilized DNA containing a site-specific psoralen adduct for in vitro transcription experiments. As shown in Fig. 3, lane 2, transcription on the immobilized DNA template showed a complete stop at the triplex site, and a very little amount of runoff transcripts was observed. Although these runoff transcripts may result from transcriptional read-through past the site of DNA damage, they could also indicate a low level of undamaged template in the preparation. It is also likely that psoralen probe can crosslink to the template at nonspecific sites not involved in transcription inhibition, which resulted in immobilization of the template and gave a small amount of runoff transcripts. To distinguish between arrested and terminated RNA pol II complexes, transcription was performed on the immobilized template, and beads were separated from solution, and washed with buffer D and stop solution to remove bound proteins. The solution phase contains terminated RNA pol II complexes (Fig. 3, lane 3), and beads carry the arrested RNA pol II complexes at the damage site (Fig. 3, lane 5). Washing the beads with buffer did not release the stalled RNA pol II complexes (Fig. 3, lane 4). It is important to note that no other transcripts were observed with the arrested RNA pol II (Fig. 3, lane 5). Our results demonstrate that a homogeneous population of elongating RNA pol II complexes can be arrested at a DNA damage site in the template produced by a psoralen adduct.
Arrest and Termination of RNA Pol II Elongation Complexes Depend upon the Nature of DNA Damage.
To determine the effect of psoralen adducts and the length of triplex probes on transcriptional termination, we designed three psoralen-containing third strands (P15, P14, and P17) and performed site-specific photocrosslinking reactions with the target template (Fig. 1). We reasoned that psoralen probes with varying oligonucleotide lengths would position psoralen at different sites in the target DNA sequence, and therefore, would result in the formation of monoadducts or crosslinks (Fig. 1B). P15 is a 15-oligonucleotide probe, and psoralen photochemistry can give rise to an interstrand crosslink between T1 and T2 nucleotides in the triplex target as well as monoadducts (11, 15). Our model studies showed that P15 gave a T1 and T2 crosslink as a major product and monoadducts at T1 and T2 sites as minor products (Y.-H. Ping and T.M.R., unpublished results). P14 is a one-base shorter probe than P15, and it gives monoadduct at T2 as a major product and interstrand crosslinks between T1 and T2 bases with lower yields. Our third probe, P17, contains two more nucleotides than P15, and it only formed a T1 and T2 interstrand crosslink and very few monoadducts (Y.-H. Ping and T.M.R., unpublished results]. After triplex formation with these three psoralen-containing third strands and photocrosslinking, in vitro transcription reactions were performed, and transcripts were analyzed on denaturing gels (Fig. 4). Because template DNA was not immobilized on magnetic beads after photocrosslinking and contains a mixture of psoralen-modified and unmodified templates, in vitro transcription reactions would yield terminated, arrested, and runoff transcripts. Therefore, the amount of runoff transcripts does not represent the RNA pol II bypassing the psoralen damage, rather it shows the presence of noncrosslinked DNA template in the transcription reaction. The major products of transcription inhibition by P15 were arrested RNA pol II elongation complexes, while termination produced minor products (Fig. 4, lane 3). In contrast to P15, P14 resulted in transcription termination as a major product, and only a small number of elongation complexes were arrested (Fig. 4, lane 4). Probe P17 effectively arrested RNA pol II elongation complexes and terminated transcription with low yields (Fig. 4, lane 5). These results show that interstrand psoralen crosslinks arrest elongating RNA pol II complexes, whereas psoralen monoadducts on the DNA template terminate transcription.
Stable Triplex Structures Terminate RNA Pol II Elongation Complexes.
We have shown that the noncrosslinked third strand cannot inhibit RNA pol II transcription elongation even at high concentration (Fig. 2). We reasoned that this could have been the result of instability of triplex formation at higher pH (8.0) conditions used for in vitro transcription experiments. Third-strand oligonucleotides conjugated with intercalators such as acridine significantly enhances the stability of triple-helical structures (16). We therefore synthesized an acridine-modified third strand Pacr (Fig. 1B) and used this to form a triplex at the target DNA sequence before performing transcription. As shown in Fig. 4 (lane 2), Pacr effectively terminated transcription at the triplex site. Our results indicate that an acridine-conjugated third strand forms a stable triple-helix, which can terminate elongating RNA pol II complexes in vitro.
Elongation Complexes Arrested at DNA Damage Site Contain RNA Pol II.
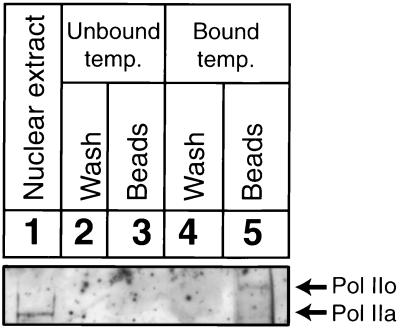
To demonstrate that RNA pol II is a part of the arrested elongation complex, site-specific DNA damage was introduced by P15 in the DNA templates and immobilized on magnetic beads. After performing in vitro transcription, arrested RNA pol II complexes were separated from solution, washed extensively with buffer D, and analyzed on SDS gels by immunoblotting (Fig. 5). Mammalian cells contain two forms of RNA pol II, phosphorylated (IIo) and nonphosphorylated (IIa), that differ in the extent of phosphorylation within the C-terminal domain of their largest subunit (17). The nonphosphorylated form of RNA pol II preferentially associates with the preinitiation complex, whereas RNA pol II derived from isolated ternary complexes is highly phosphorylated (18, 19). In HeLa cell extracts (depending upon the method of preparation), the majority of the RNA pol II is nonphosphorylated, and all of the RNA pol II detected in a stalled elongation complex is phosphorylated (12). In agreement with previous reports (12, 17, 19), phosphorylated RNA pol II (IIo) was detected in the arrested elongation complexes (Fig. 5, lane 5). This observation supported the identity of the arrested complexes as valid elongation complexes. Extensive wash of beads did not release any detectable amount of bound RNA pol II (Fig. 5, lane 4). Control experiments showed that magnetic beads did not bind RNA pol II when transcription reactions were carried out on unbound templates (Fig. 5, lane 3). Stability of arrested RNA pol II complexes was tested by incubating RNA pol II containing beads at room temperature for 0.5 to 3 hr followed by analysis of released RNA transcripts (data not shown). RNA pol II complexes isolated on the beads were stable at least for 3 hr at room temperature, and no released RNA transcripts were detected. Our results demonstrate that arrested elongation complexes are very stable and contain nascent RNA transcripts and phosphorylated forms (IIo) of RNA pol II.
Figure 5.
Visualization of arrested RNA pol II by immunoblotting. Lane 1, 5% of the nuclear extract present in the transcription reaction. Transcription was performed with unbound template in the presence of beads: lane 2, buffer D wash of beads; lane 3, beads treated with SDS loading buffer. P15 (10 μM) was used to form triplex and UV irradiated, and this template was immobilized on magnetic beads and used for transcription. Bound RNA pol II complexes were separated by magnet: buffer D wash (lane 4) and beads treated with SDS loading buffer (lane 5). RNA pol II (Pol IIa, unphosphorylated form; Pol IIo, phosphorylated form) is shown.
Mapping the Arrest and Termination Sites.
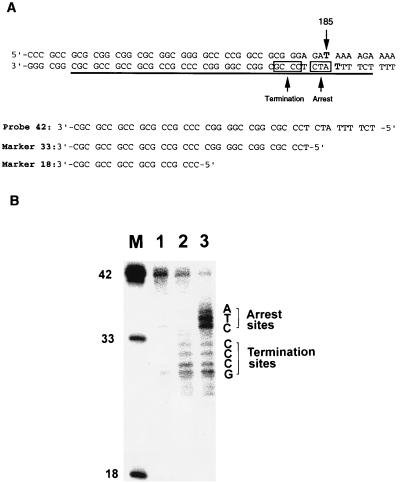
To determine the effect of triplex structure and psoralen damage on transcription, we mapped the 3′-end of RNA transcripts by nuclease S1 analysis. We synthesized an S1 probe, P42, covering the region 150–191 in the template DNA, which includes 36 base pairs upstream from the psoralen adduct site and six base pairs within the triplex site (Fig. 6A). After isolating the transcripts and hybridizing to the P42 probe, nuclease S1 digestion was performed, and the protected probes were analyzed on denaturing gels (20). The length of protected probes was determined by using oligonucleotide markers of known sequences (Fig. 6A). Results of this analysis are shown in Fig. 6. Because a stable triple-helix terminates transcription, RNA transcripts arising from only termination of elongation were generated by using Pacr probe to form a stable triple-helix before performing transcription reactions (Fig. 4). These termination RNA products were hybridized to P42 probe and subjected to S1 reaction. Nuclease S1 digestion of P42 probe labeled at its 3′-end gave a number of DNA fragments containing 29–32 nucleotides, indicating that termination leads to a population of RNA transcripts ranging in length from 178 to 181 nucleotides (Fig. 6B, lane 2). Psoralen probe, P15, was used to introduce psoralen damage in the DNA template, and in vitro transcription was performed without isolation of unmodified template. RNA transcripts originating from unmodified and psoralen crosslinked DNA templates were isolated, hybridized to S1 probe, and digested with S1 nuclease. Two populations of digested DNA fragments were observed: 29–32 nucleotides corresponding to termination and 34–36 nucleotides with high intensity bands representing the arrest of RNA pol II. Because P15 causes both transcriptional arrest and termination (Fig. 4), one would expect to obtain two populations of RNA transcripts from DNA templates damaged with P15 photocrosslinking. The intensity of 34–36 nucleotide bands is higher than the termination products because P15 crosslinking reaction yields more arrest than termination of elongation (Fig. 6, lane 3). Undigested probe is the result of hybridization of P42 to the full-length runoff transcripts, which are 340 nucleotides long. These results indicate that RNA transcripts resulting from RNA pol II arrest at the psoralen adduct site contain 183–185 nucleotides. As a control experiment, full-length (340 nucleotides) runoff transcripts were prepared by using unmodified templates. After hybridizing to the S1 probe, nuclease digestion of the full-length runoff transcripts did not give any smaller fragments obtained by other RNA transcripts (Fig. 6, lane 1). These results show that elongating RNA pol II complex can transcribe up to the psoralen adduct site on the template strand, however, it also gets arrested one or two nucleotides before the adduct site.
Figure 6.
S1 mapping analysis of the 3′-end of RNA transcripts produced by transcriptional arrest and termination. (A) Sequences of the template DNA near the region of triplex site, S1 probe (probe 42), marker 33, and marker 18 are shown. Location of the probe 42 is underlined in the template DNA. Transcriptional arrest and termination sites are shown in two square boxes. Psoralen crosslink site at 185 is indicated by an arrow, and two T residues involved in crosslink formation are highlighted. (B) Analysis of the reaction products of an S1 nuclease mapping experiment. RNA transcripts were hybridized to probe 42 labeled at its 3′-end with 32P and digested with S1 nuclease, and DNA fragments were analyzed on a sequencing gel. Lane 1, a full-length (340 nucleotides) runoff transcript prepared by using unmodified template. Lane 2, RNA transcripts arising from only termination of elongation were generated by using Pacr probe to form a stable triple helix before performing in vitro transcription reactions. Lane 3, RNA transcripts produced from DNA templates damaged with psoralen adducts. Psoralen probe, P15, was used to form psoralen adducts in the DNA template and in vitro transcription was performed without isolation of unmodified template. Lane M, marker lane containing probe 42, marker 33, and marker 18 oligonucleotides. Sequences of termination and arrest sites are labeled.
DISCUSSION
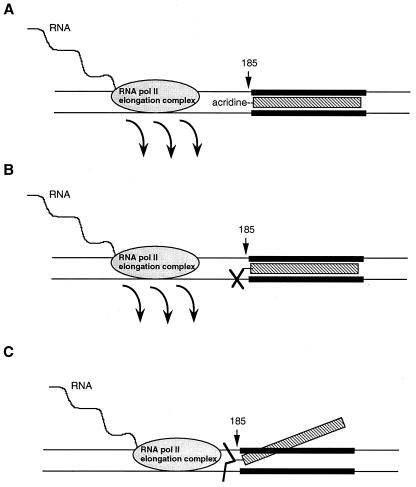
We have developed a new method to study the effect of DNA damage and triplex structure in DNA templates on RNA pol II elongation. A summary of our findings is presented in the model shown in Fig. 7. RNA pol II elongation complex is arrested by a psoralen interstrand crosslink in the template DNA and transcribes RNA until it reaches the damage site. However, most of the RNA pol II elongation complexes stall 1–2 nucleotides before the psoralen crosslink site. On the other hand, psoralen monoadducts and a stable triple-helical DNA terminate transcription. This termination occurs 4–8 nucleotides before RNA pol II elongation complex reaches the psoralen adduct site or the triple-helix in the template (Fig. 7 A and B). These results establish that a psoralen crosslink and a triple-helical DNA structure are absolute barriers to RNA pol II transcription elongation. Hearst and coworkers reported that elongating T7 and Escherichia coli RNA polymerases were blocked by a site-specifically placed psoralen diadduct (5–8). Our results show that an RNA pol II elongation complex is arrested by an interstrand psoralen crosslink, similar to T7 and Escherichia coli RNA polymerases. However, we observed efficient RNA pol II arrest 1–2 nucleotides before the psoralen-adduct site, an observation different from T7 and E. coli polymerases. The reasons for this difference are not clear at this point, however, three possible explanations can be put forward: (i) due to the larger size of RNA pol II elongation complex, it usually cannot transcribe up to the crosslink site and gets stalled 1–2 nucleotides earlier while the other two smaller polymerases can continue transcription up to the crosslink site; (ii) the presence of third strand near the psoralen crosslink site forms a different DNA structure than the structure used in E. coli and T7 RNA polymerases experiments, and elongating RNA pol II stops 1–2 nucleotides earlier than other polymerases due to the steric hindrance of third strand; and (iii) another possibility cannot be ruled out that the arrest of RNA pol II elongation machinery at a DNA damage site involves a more complex regulatory mechanism responsible for the difference in behavior of RNA pol II arrest from other RNA polymerases.
Figure 7.
Model showing the effect of DNA damage on transcription elongation. (A) A stable triplex structure is difficult to melt during elongation by RNA pol II and destabilizes the elongation complex. This leads to termination 4–8 nucleotides before the elongation complex encounters triplex. (B) Psoralen monoadduct covalently staples the third strand to the double helix without significantly destabilizing the triplex structure. Elongating RNA pol II cannot unwind the triplex, therefore, it leaves the template 4–8 nucleotides before reaching the damage site. (C) An interstrand psoralen crosslink contracts the major groove and disrupts the triplex structure. RNA pol II elongation complex can unwind this DNA template and transcribes RNA until it reaches psoralen crosslink site, at that point RNA pol II gets arrested and waits for DNA damage repair.
What is the effect of DNA structure on transcription elongation? Triple-helical DNA is formed by laying a third strand into the major groove of DNA (21). The third strand of a triplex forms (Hoogsteen) hydrogen bonds with the available functional groups of each base pair in a Watson–Crick double-helix. Recent studies have shown that stable triplex structures act as repressors and inhibitors of transcription (22–24). Elongation complexes are stable at nonterminator positions and intrinsic termination sites could destabilize these complexes (25, 26). We show here that a stable triplex DNA provides a structure that is difficult to melt during transcription and destabilizes elongation complexes, leading to termination. Triplex structure is stabilized by using either an acridine-modified third strand or attaching it to the target double-helix by a psoralen monoadduct. In the case of interstrand psoralen crosslink, elongation complexes were arrested and not terminated, suggesting that a psoralen diadduct destabilizes triplex structure. Further support for this model comes from recent NMR studies of psoralen-adducted DNA. Spielmann et al. (27, 28) reported solution structures of psoralen monoadducted and crosslinked DNA oligomers determined by NMR spectroscopy and restrained molecular dynamics. Helical parameters for the monoadducted and crosslinked DNA oligomers were calculated and compared with canonical A-DNA and B-DNA. Analysis of the helical parameters revealed that formation of the crosslink causes a contraction across the major groove and widens and makes shallower the minor groove at the psoralen adduct site. A-form DNA has a deep major groove and a wide, shallow minor groove, while B-form DNA has a wide major groove and a relatively narrow deep minor groove. As the DNA alters its conformation to accommodate the psoralen photoadducts, the crosslink and to a lesser extent, monoadducts, take on the appearance of an A-form helix (27, 28). Because the third strand of a triplex DNA occupies the major groove of double-helix, we propose that an interstrand psoralen crosslink disrupts triplex DNA by contracting the major groove and destabilizing the (Hoogsteen) hydrogen bonds. This disruption of the triplex may bring DNA structure back to a duplex-like DNA. Psoralen monoadduct contracts major groove to a lesser extent and does not destabilize a triplex. In light of the above studies, we suggest that a stable triplex DNA acts as a termination site for transcription where RNA pol II complex is destabilized and leaves the template a few nucleotides before reaching the triplex structure (Fig. 7). In the case of interstrand psoralen crosslink, the triplex is less stable or disrupted, and the elongation complex keeps unwinding the template DNA until it approaches the damage site and gets arrested waiting for DNA damage repair.
RNA pol II is a dynamic molecule and undergoes continued conformational transitions during transcription elongation (29). During transcription elongation, template DNA sequences can lead RNA pol II to pause, become arrested, terminate, or pass through terminators. DNA damage in the templates can stall in vitro reconstituted RNA pol II elongation complexes (30, 31). In this study, we used HeLa nuclear extract to carry out in vitro transcription. Our results describe the first report, to our knowledge, showing that DNA damage can arrest RNA pol II elongation complexes from HeLa nuclear extracts. This finding is very intriguing because HeLa nuclear extracts contain DNA damage repair and other regulatory proteins that apparently cannot disengage RNA pol II elongation complexes stalled at an interstrand psoralen crosslink site in DNA template. Therefore, it suggests the possibility that RNA pol II elongation complex arrested at a damage site does not dissociate from the template while DNA damage is being repaired.
The method of delivering site-specific psoralen damage by a triplex structure and isolation of arrested RNA pol II elongation complexes should be generalizable to any promoter and DNA template sequences. This strategy provides a new approach to study the mechanism of transcription elongation and transcription-coupled DNA damage repair, and to purify DNA damage repair proteins.
Acknowledgments
We thank Nicholas Keen and Jonathan Karn for generous gifts of HIV-1 long terminal repeat containing plasmids. Special thanks to E. Peter Geiduschek, John E. Hearst, and Peter H. von Hippel for critical reading of the manuscript and valuable suggestions. We are also grateful to Beate Schwer and Alexey Ryazanov for helpful discussions, and to Xilu Wang for the synthesis of psoralen phosphoramidites. This work was supported in part by the National Institutes of Health Grant AI 34785 and by the State of New Jersey Commission on Cancer Research. T.M.R. is a recipient of a Research Career Development Award from the National Institutes of Health.
ABBREVIATION
- RNA pol II
RNA polymerase II
References
- 1.Kerppola T K, Kane C M. FASEB J. 1991;5:2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- 2.Jones K A, Peterlin B M. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 3.Reines D, Conaway J W, Conaway R C. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 4.Rice G A, Chamberlin M J, Kane C M. Nucleic Acids Res. 1993;21:113–118. doi: 10.1093/nar/21.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y-B, Gamper H, Houten B V, Hearst J E. J Mol Biol. 1988;199:277–293. doi: 10.1016/0022-2836(88)90314-2. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y-B, Gamper H, Hearst J E. J Biol Chem. 1988;263:527–534. [PubMed] [Google Scholar]
- 7.Sastry S S, Hearst J E. J Mol Biol. 1991;221:1111–1125. [PubMed] [Google Scholar]
- 8.Sastry S S, Hearst J E. J Mol Biol. 1991;221:1091–1110. [PubMed] [Google Scholar]
- 9.Krummel B, Chamberlin M J. J Mol Biol. 1992;225:239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- 10.Krummel B, Chamberlin M J. J Mol Biol. 1992;225:221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- 11.Takasugi M, Guendouz A, Chassingnol M, Decout J L, Lhomme J, Thuong N T, Hélène C. Proc Natl Acad Sci USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keen N J, Gait M J, Karn J. Proc Natl Acad Sci USA. 1996;93:2505–2510. doi: 10.1073/pnas.93.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deissler H, Behn-Krappa A, Doerfler W. J Biol Chem. 1996;271:4327–4334. doi: 10.1074/jbc.271.8.4327. [DOI] [PubMed] [Google Scholar]
- 15.Vasquez K M, Wensel T G, Hogan M E, Wilson J W. Biochemistry. 1996;35:10712–10719. doi: 10.1021/bi960881f. [DOI] [PubMed] [Google Scholar]
- 16.Zhou B-W, Puga E, Sun J-S, Garestier T, Hélène C. J Am Chem Soc. 1995;117:10425–10428. [Google Scholar]
- 17.Dahmus M E. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 18.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zawel L, Kumar K P, Reinberg D. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 7.66–7.70. [Google Scholar]
- 21.Vlieghe D, Meervelt L V, Dautant A, Gallois B, Précigous G, Kennard O. Science. 1996;273:1702–1705. doi: 10.1126/science.273.5282.1702. [DOI] [PubMed] [Google Scholar]
- 22.Grigoriev M, Praseuth D, Guieysse A L, Robin P, Thuong N T, Hélène C, Harel-Bellan A. Proc Natl Acad Sci USA. 1993;90:3501–3505. doi: 10.1073/pnas.90.8.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giovannangeli C, Perrouault L, Escudé C, Thuong N, Hélène C. Biochemistry. 1996;35:10539–10548. doi: 10.1021/bi952993x. [DOI] [PubMed] [Google Scholar]
- 24.Giovannangeli C, Perrouault L, Escudé C, Gryaznov S, Hélène C. J Mol Biol. 1996;261:386–398. doi: 10.1006/jmbi.1996.0471. [DOI] [PubMed] [Google Scholar]
- 25.von Hippel P H, Yager T D. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 26.von Hippel P H, Rees W A, Rippe K, Wilson K S. Biophys Chem. 1996;59:231–246. doi: 10.1016/0301-4622(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 27.Spielmann H P, Dwyer T J, Sastry S S, Hearst J E, Wemmer D E. Proc Natl Acad Sci USA. 1995;92:2345–2349. doi: 10.1073/pnas.92.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spielmann H P, Dwyer T J, Hearst J E, Wemmer D E. Biochemistry. 1995;34:12937–12953. [PubMed] [Google Scholar]
- 29.Chamberlin M J. Harvey Lect. 1992;88:1–21. [PubMed] [Google Scholar]
- 30.Donahue B A, Yin S, Taylor J-S, Reines D, Hanawalt P C. Proc Natl Acad Sci USA. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donahue B A, Fuchs R P P, Reines D, Hanawalt P C. J Biol Chem. 1996;271:10588–10594. doi: 10.1074/jbc.271.18.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]