Abstract
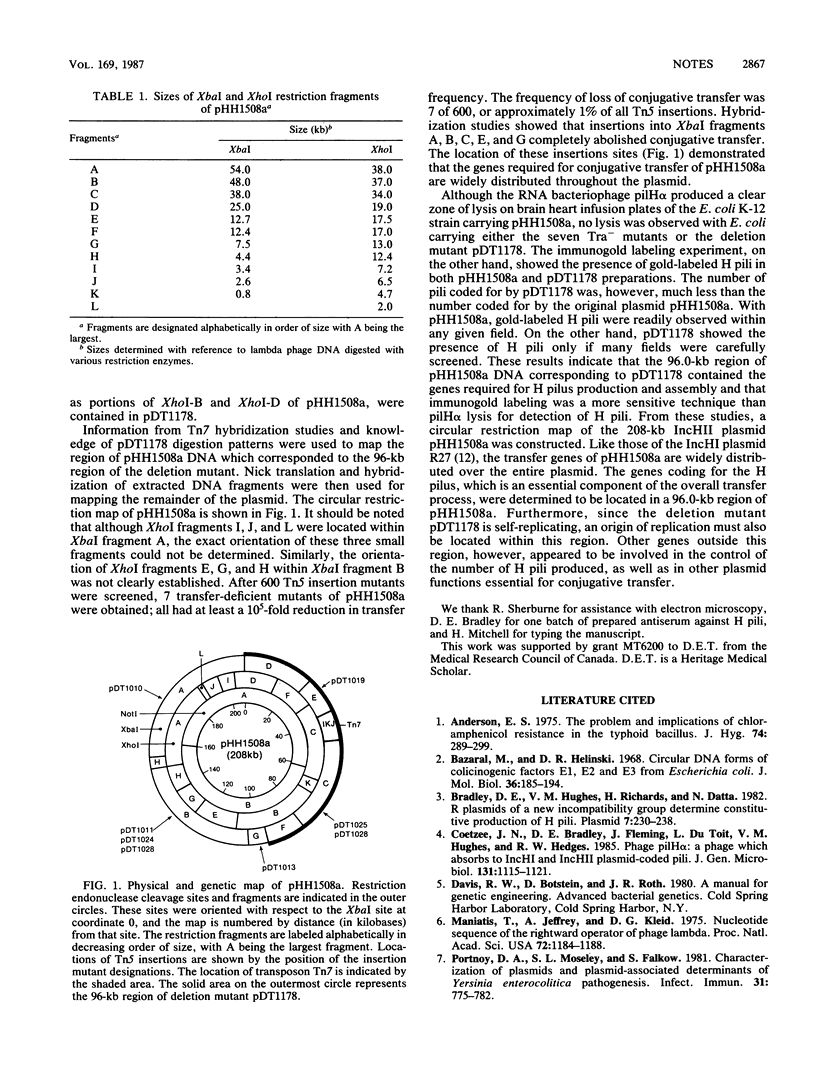
A circular map of the 208-kilobase IncHII plasmid pHH1508a was constructed by using the restriction endonucleases XbaI, XhoI, and NotI. Tn5 insertion mutagenesis showed that transfer genes were distributed widely over the plasmid map. Analysis of the deletion mutant pDT1178 indicated that genes required for conjugative pilus production are located in a 96.0-kilobase region of the plasmid.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S. The problem and implications of chloramphenicol resistance in the typhoid bacillus. J Hyg (Lond) 1975 Apr;74(2):289–299. doi: 10.1017/s0022172400024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Hughes V. M., Richards H., Datta N. R plasmids of a new incompatibility group determine constitutive production of H pili. Plasmid. 1982 May;7(3):230–238. doi: 10.1016/0147-619x(82)90004-x. [DOI] [PubMed] [Google Scholar]
- Coetzee J. N., Bradley D. E., Fleming J., du Toit L., Hughes V. M., Hedges R. W. Phage pilH alpha: a phage which adsorbs to IncHI and IncHII plasmid-coded pili. J Gen Microbiol. 1985 May;131(5):1115–1121. doi: 10.1099/00221287-131-5-1115. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy D. A., Moseley S. L., Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981 Feb;31(2):775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. W. Letter: Thermosensitive transfer factors in chloramphenicol-resistant strains of Salmonella typhi. Lancet. 1974 Aug 3;2(7875):281–282. doi: 10.1016/s0140-6736(74)91435-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Taylor D. E., Brose E. C., Kwan S., Yan W. Mapping of transfer regions within incompatibility group HI plasmid R27. J Bacteriol. 1985 Jun;162(3):1221–1226. doi: 10.1128/jb.162.3.1221-1226.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E., Brose E. C. Restriction endonuclease mapping of R27 (TP117), an incompatibility group HI subgroup 1 plasmid from Salmonella typhimurium. Plasmid. 1985 Jan;13(1):75–77. doi: 10.1016/0147-619x(85)90058-7. [DOI] [PubMed] [Google Scholar]
- Taylor D. E. Transfer-defective and tetracycline-sensitive mutants of the incompatibility group HI plasmid R27 generated by insertion of transposon 7. Plasmid. 1983 May;9(3):227–239. doi: 10.1016/0147-619x(83)90001-x. [DOI] [PubMed] [Google Scholar]
- Whiteley M., Taylor D. E. Identification of DNA homologies among H incompatibility group plasmids by restriction enzyme digestion and Southern transfer hybridization. Antimicrob Agents Chemother. 1983 Aug;24(2):194–200. doi: 10.1128/aac.24.2.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobec E. A., Frost L. S., Pieroni P., Armstrong G. D., Hodges R. S., Parker J. M., Finlay B. B., Paranchych W. Location of the antigenic determinants of conjugative F-like pili. J Bacteriol. 1986 Aug;167(2):660–665. doi: 10.1128/jb.167.2.660-665.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]



