Abstract
Structural studies of authentic HIV reverse transcriptase (RT) suggest a role for the p51 carboxyl terminus in forming an active RNase H conformation [Rodgers, D. W., Gamblin, S. J., Harris, B. A., Ray, S., Culp, J. S., Hellmig, B., Woolf, D. J., Debouck, C. & Harrison, S. C. (1995) Proc. Natl. Acad. Sci. USA 92, 1222–1226]. We have purified mutant RT heterodimers containing deletion of 5, 9, or 13 amino acids from the p51 carboxyl terminus. These “selectively deleted” heterodimers have been analyzed for changes in RNA-dependent DNA polymerase activity, RNase H activity, and the ability to catalyze DNA strand transfer. As deletions extended into the p51 subunit, a decrease in the stability of the RT–DNA complex was apparent. The largest effect was observed for p66/p51Δ13 RT, which showed a 3-fold decrease relative to wild-type RT. RNase H activity was measured by digestion of the RNA in a 5′ 32P-labeled RNA/DNA hybrid. Deletion of 5 or 9 amino acids from p51 had little effect on synthesis-dependent and synthesis-independent RNase H activities. In contrast, deletion of 13 amino acids from p51 increased the length of the hydrolysis products of both RNase H activities by 8–10 bp, thus changing the spatial relationship between the polymerase and RNase H active sites from a distance of 17–18 bp to 26–27 bp. The Δ13 derivative was also incapable of efficient DNA strand transfer. This defect in strand transfer could be suppressed by the 71-amino acid form of HIV nucleocapsid protein (NC) but not by the 55-amino acid form (NC55) or by equine infectious anemia virus NC. These results provide evidence for the existence of a specific complex between RT and NC and are discussed in terms of the role of this complex in proviral DNA synthesis.
Human immunodeficiency virus (HIV) is the causative agent of acquired immune deficiency syndrome (AIDS) (1, 2). As with all retroviruses, establishment of HIV infection requires integration of double-stranded viral DNA into the host genome (3). Synthesis of viral DNA from genomic RNA is catalyzed by the virus-encoded RNA/DNA-dependent DNA polymerase, reverse transcriptase (RT). Inhibitors of RT activity or DNA elongation are therefore effective antiviral agents (4). Current models of RT-catalyzed viral DNA synthesis require the enzyme to complete two strand transfer reactions (template switches) (5). The first strand transfer reaction permits strong-stop, minus-strand DNA to be used as primer for completion of full-length, minus-strand DNA. Studies of this reaction on model substrates have indicated that, in addition to the DNA polymerase activity of RT, two kinetically distinct RNase H activities of RT are also required (6, 7).
The polymerase-dependent RNase H activity associated with RT makes endonucleolytic cleavages in the RNA 17–18 nt behind the 3′ terminus of nascent DNA (7). This 17- to 18-nt distance is dictated by the spatial separation between the catalytic sites of RT (7). Moreover, the rate of RNA hydrolysis relative to nucleotide incorporation is sufficiently slow that, upon completion of strong-stop DNA synthesis, RNA oligonucleotides remain bound to the newly formed DNA product (6, 7). Hydrolysis of these RNA oligonucleotides from a length of 18 nt to ≈9 nt is the function of the synthesis-independent RNase H activity of RT (6, 7) and causes a lag in the accumulation of product during the strand transfer reaction in vitro (6). RNA oligonucleotides of 9 nt (or shorter) most likely dissociate from the strong-stop DNA (6). Interestingly, the synthesis-independent RNase H activity of HIV RT is stimulated by the presence of HIV nucleocapsid protein (NC) (8).
In this study, we have taken advantage of one of the high resolution crystal structures for HIV RT (9) to identify residues of this enzyme that might be important in determining the fixed distance between the two catalytic sites of the RT heterodimer. We find that deletion of 5, 9, or 13 amino acids from the carboxyl terminus of the p51 subunit of RT produces heterodimeric RTs with polymerase and RNase H activities essentially indistinguishable from the wild-type enzyme. However, deletion of 13 amino acids from the p51 carboxyl terminus changes the spatial relationship between the polymerase and RNase H active sites from a distance equivalent to 17–18 bp to 26–27 bp. This change also reduces the efficiency of strand transfer. The specific defect in strand transfer appears to reflect the inability of synthesis-independent RNase H activity of this mutant to produce RNA hydrolysis products capable of dissociating from strong-stop DNA. This defect, however, can be suppressed by the presence of the 71 amino acid form of HIV NC but not by the 55 amino acid form (NC55) or by equine infectious anemia virus (EIAV) NC. These data support the existence of a virus-specific RT–NC complex and are discussed in terms of the role of this complex during viral DNA synthesis.
MATERIALS AND METHODS
DNA oligonucleotides were purchased from Operon Technologies (Alameda, CA). Ultrapure dideoxythymidine triphosphate (ddTTP) deoxyribonucleotides (dNTPs), ribonucleotides, oligo-dT16, and poly(A) were from Pharmacia. [γ-32P]ATP was from DuPont/NEN. All additional reagents were molecular biology grade from Sigma. T4 polynucleotide kinase and calf intestinal alkaline phosphatase were from Amersham. Wild-type and mutant RTs were constructed and purified as described (10, 11). Purified HIV NC was a gift of David Giedroc (Texas A&M University). Purified HIV NC55 was a gift of Robert Gorelick (SAIC-Frederick, Frederick Cancer Research Center). Purified EIAV NC was a gift of Ronald Montelaro (University of Pittsburgh).
Oligonucleotide Substrates.
RNA templates were synthesized by runoff transcription with T7 RNA polymerase and purified by denaturing PAGE as described (6–8). DNA oligonucleotides were 5′ 32P-labeled following the manufacturer’s protocol using T4 polynucleotide kinase. Prior to labeling RNA, terminal phosphates were removed using calf intestinal alkaline phosphatase. Duplexes were prepared such that the concentration of unlabeled nucleic acid was 10% greater than that of the labeled nucleic acid. Annealing reactions were completed in 50 mM Tris⋅HCl (pH 8.0). Nucleic acids were heated to 70°C for 5 min followed by slow cooling to 25°C. Integrity of the duplexes was verified by native PAGE. The oligonucleotide sequences are as follows: 20-nt DNA primer, 5′-AGAGCTCCCAGGCTCAGATC-3′; 40-nt primary RNA template, 3′-UCUCGAGGGUCCGAGUCUAGACCAGAUUGGUCUCUCUCUGGG-5′; 41-base RNA acceptor template, 3′-ACCAGAUUGGUCUCUCUGGGUCAUGUCCGUUUUUCGUCGGAG-5′; 40-nt DNA complement to primary RNA template, 5′-AGAGCTCCCAGGCTCAGATCTGGTCTAACCAGAGAGACCC-3′.
All reactions containing wild-type and mutant RTs were performed at 37°C in a final volume of 50 μl containing 7 mM MgCl2, 0.1 mM dNTPs (when required), 0.1 mM ddTTP (when required), 50 mM Tris⋅HCl (pH 8.0), 75 mM KCl, 1 mM DTT, and 0.1% Triton X-100. Reaction products were resolved from substrates by denaturing PAGE (see Fig. 3). Products and substrates were visualized and quantified by phosphorimaging using the imagequant software provided with the Molecular Dynamics PhosphorImager.
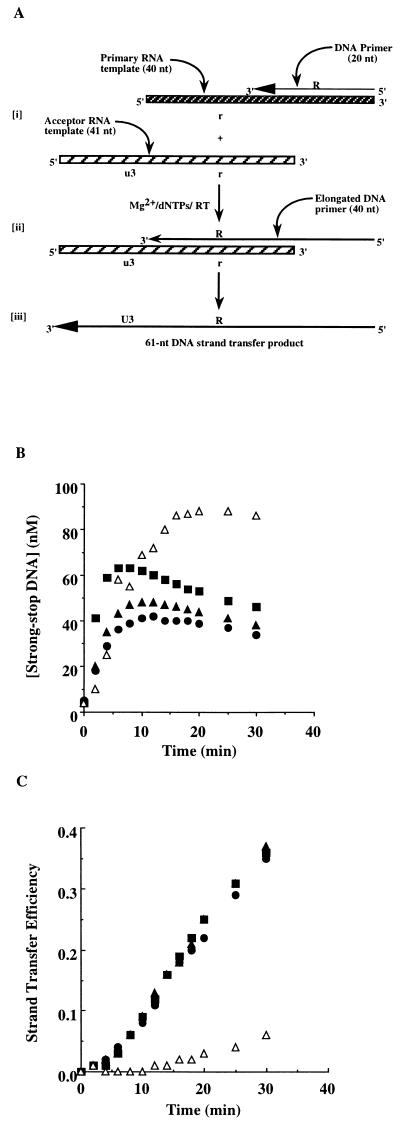
Figure 3.
Strand transfer reactions catalyzed by selectively deleted RTs. (A) Schematic representation of the strand transfer assay. [i] A primer/template, comprising a 20-nt DNA and a 40-nt RNA, is incubated with RT in the presence of divalent cation and deoxynucleotides as described. [ii] This produces a 40-nt strong-stop DNA which is transferred to the acceptor template. [iii] Continued synthesis produces a 61-nt DNA strand transfer product. (B) Time course of strong-stop DNA synthesis by wild-type (•), p66/p51Δ5 (▴), p66/p51Δ9 (▪), and p66/p51Δ13 (▵) RTs. (C) Time course of the strand transfer efficiency (i.e., the concentration of 61-nt DNA relative to the sum of 40-nt and 61-nt DNAs) of wild-type (•), p66/p51Δ5 (▴), p66/p51Δ9 (▪), and p66/p51Δ13 (▵) RTs.
Polymerase Assay.
Varying concentrations of RT (0–100 nM), based on a molar extinction coefficient at 280 nm of 260,450 (12), were preincubated with 1 μM oligo-dT16/poly(A) for 5 min; the reaction was initiated by addition of MgCl2 and ddTTP. Samples (5 μl) were quenched by dilution into 20 μl of 0.5 M EDTA at 1-min intervals over a 5-min total reaction time. Product concentration was plotted as a function of time and the data fit to a line. The intercept indicated the concentration of active enzyme in the reaction, and the slope indicated the rate of single nucleotide incorporation as reported in Table 1.
Table 1.
Comparison of polymerase and RNase H activities for wild-type (WT) and mutant RTs
| Enzyme | Active enzyme, % | Polymerase activity, min−1 | RNase H activity, min−1 |
|---|---|---|---|
| WT | 56 ± 3 | 0.86 ± 0.05 | 1.04 ± 0.10 |
| Δ5 | 52 ± 11 | 1.14 ± 0.07 | 2.70 ± 0.50 |
| Δ9 | 28 ± 8 | 2.28 ± 0.09 | 3.10 ± 0.50 |
| Δ13 | 42 ± 5 | 2.67 ± 0.13 | 2.50 ± 0.30 |
RNase H Assays.
Assays to determine the steady-state rate of RNA hydrolysis were performed using a duplex composed of a labeled 40-nt RNA primary template and its 40-nt DNA complement. Various concentrations of RT (0–100 nM) were preincubated with duplex (1 μM) for 5 min, and the reactions were initiated by the addition of MgCl2. Aliquots were quenched as described above at 15-sec intervals over a 2-min total reaction time. Product concentration was plotted as a function of time and the data fit to a line. The slope provided the rate of RNA hydrolysis reported in Table 1.
Alternatively, RNase H activity was determined using a 90-nt 5′ 32P-labeled RNA template/36-nt DNA primer sequence. The oligonucleotide sequences are as follows: 90-nt RNA template, 5′-GGGCGAAUUCGAGCUCGGUACCCGAUAGGGACUAGGCCCUUCUAAUUCCCCUUUCCCCCUCUCCUGGUGAUCCUUUCCAUCCCUGUCCCC-3′; 36-nt DNA primer, 3′-GGGGGAGAGGACCACTAGGAAAGGTAGGGACAGGGG-5′. Hydrolysis was monitored in the absence of DNA synthesis. Samples containing a range of enzyme concentrations were analyzed after a 5-sec incubation period with the RNA–DNA hybrid. Hydrolysis products were evaluated by high voltage denaturing electrophoresis followed by autoradiography. Product lengths were determined by co-electrophoresis of DNA markers.
Strand Transfer Assay.
RT (100 nM) was preincubated with a primer/template (100 nM) composed of the 20-nt DNA primer hybridized to the 40-base primary RNA template in the presence of the acceptor RNA (240 nM) for 5 min. The reaction was initiated by the addition of MgCl2 and dNTPs. Samples were quenched as described above at various intervals over a 20-min total reaction time. When NC was included in the reaction, it was present during the initial preincubation. Concentrations used are indicated in appropriate figure legends.
RESULTS AND DISCUSSION
A Role for the Carboxyl Terminus of the p51 Subunit of RT in Maintaining the Active Conformation of RNase H.
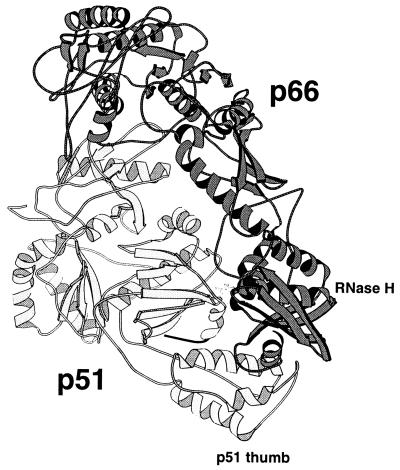
Based on the structure of HIV RT shown in Fig. 1, it appears that the compact p51 subunit stabilizes the more extended conformation of p66. Interaction with the p51 thumb subdomain, in particular, stabilizes the position of the RNase H domain of p66, thereby defining the spatial relationship between the polymerase and RNase H active sites. The p51 thumb itself, however, is positioned primarily by interactions of its carboxyl-terminal helix. Therefore, deletion of this helix might alter the position of both the p51 thumb and the RNase H domain, changing the spatial relationship between the two active sites. To test this hypothesis, RTs were constructed which had 5, 9, or 13 amino acids removed from the carboxyl terminus of p51. Deleted p51 subunits were reconstituted with wild-type p66 subunits, and these “selectively deleted” heterodimers were purified to homogeneity as described (10, 11).
Figure 1.
Structure of HIV RT. The p66 subunit of unliganded RT is darkly shaded, and the p51 subunit is light except for the carboxyl-terminal residues (426–434) drawn in black (9). Six residues (435–440) at the extreme carboxyl terminus of p51 were not present in the crystal structure and are not shown. The RNase H domain of p66 and the thumb domain of p51 are indicated. The drawing was produced with the program molscript.
Selectively Deleted Heterodimers Retain Polymerase and RNase H Activities Comparable to Wild-Type RT.
The steady-state rate of ddTTP incorporation of each purified, RT derivative was measured as described. As indicated in Table 1, ≈50% of each RT preparation was active and is similar to the value observed for the wild-type enzyme, consistent with the previous report by Kati et al. (12). However, the rate of dissociation of each RT derivative from primer/template was increased ≈3-fold (Table 1). The steady-state rate of RNA hydrolysis was measured as described. This assay is used only to demonstrate catalytic activity of the RNase H domain independent of spatial and temporal constraints imposed by polymerization. As indicated in Table 1, the steady-state rate of RNA hydrolysis for the RTs is comparable to that measured for polymerase activity, demonstrating that each RT retains RNase H activity. Furthermore, this result suggests that access of substrate to the RNase H active site and stability therein are dictated by the same interactions required for polymerization, providing evidence for conservation in the overall organization of the altered heterodimers relative to that of wild-type RT.
Deletion of 13 Amino Acids from the Carboxyl Terminus of p51 Alters the Spatial Relationship Between the Polymerase and RNase H Active Sites.
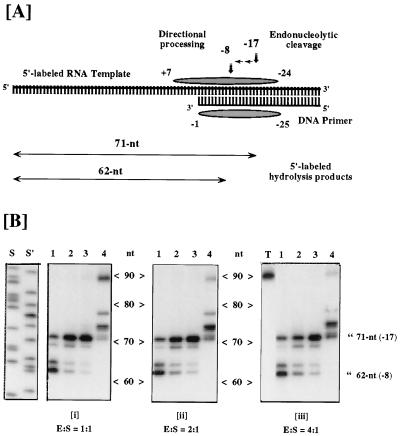
To determine whether the carboxyl-terminal helix of p51 is important for maintaining the 17- to 18-bp separation of the polymerase and RNase H active sites, an experiment was performed using the substrate shown in Fig. 2A. Incubating wild-type enzyme with this substrate in the presence of magnesium results in RNase H-mediated hydrolysis of the template RNA 17–18 bp behind the 3′ terminus of the DNA primer thus producing a 71-nt RNA product (13). The first site of endonucleolytic cleavage of the RNA is indicative of the position of the RNase H active site relative to the polymerase active site which is positioned over the 3′ hydroxyl group of the DNA primer (14, 15). During a single binding event, the 71 nt RNA is the sole cleavage product observed (13). However, multiple binding events permit the directional-processing activity to trim the RNA an additional 9 nt to form a 62-nt RNA product. This product is incapable of remaining hybridized to the DNA primer owing to the instability of the 8-bp duplex between it and the primer (13).
Figure 2.
RNase H activity of selectively deleted RTs. (A) Substrate for RNase H activity is a 90-nt 5′-end labeled RNA hybridized at its 3′ terminus to a 36-nt DNA. Shaded ellipsoids represent template and primer nucleotides covered by HIV-1 RT located over the primer 3′ OH. In the absence of DNA synthesis, endoribonuclease activity ≈17 nt distal to the primer terminus produces a radiolabeled 71-nt product. Subsequent to this, a directional processing activity hydrolyzes in a 3′ → 5′ direction as far as template nucleotide −8 (62-nt product), after which the fragmented template presumably dissociates from primer. (B) RNase H activity of HIV-1 RT derivatives harboring p51 deletions. The ratio of RT to template-primer is indicated below each panel. Lanes: 1, wild-type p66/p51; 2, p66/p51Δ5; 3, p66/p51Δ9; 4, p66/p51Δ13; S and S′, DNA sequence ladders for molecular weight determination; T, no enzyme. Migration positions of the −17 and −8 hydrolysis products are indicated.
While deletion of 5 or 9 amino acids from the p51 carboxyl terminus had no effect on the site of the first RNase H-mediated cleavage, removing 13 amino acids produced an enzyme which cleaved 26–27 bp behind the 3′ terminus of the DNA primer instead of 17–18 bp (Fig. 2B). In each case the directional processing activity removed an additional 9 nt of RNA (Fig. 2B). Furthermore, even under conditions of excess enzyme and prolonged incubation, p66/p51Δ13 RT was incapable of producing a 62-nt RNA product (Fig. 2B and data not shown). These results suggest that the RNase H domain of this RT derivative is now located at ≈26–27 bp from the polymerase catalytic center, representing an increase of 8–10 bp relative to wild-type RT. These results support our hypothesis regarding the significance of the p51 carboxyl terminus for RT structure and function.
Deletion of 13 Amino Acids from the Carboxyl Terminus of p51 Impairs DNA Strand Transfer Activity.
The strand transfer assay developed by Peliska and Benkovic (6) was used to determine the effect of the deletions in p51 on this activity of the reconstituted heterodimers. The assay is illustrated in Fig. 3A. Using this assay, it is possible not only to follow the production of the strand transfer product but also to assess the efficiency of strong-stop DNA synthesis and both the synthesis-dependent and synthesis-independent RNase H activities. RNase H activities are monitored by labeling the 40-nt RNA template rather than the DNA primer.
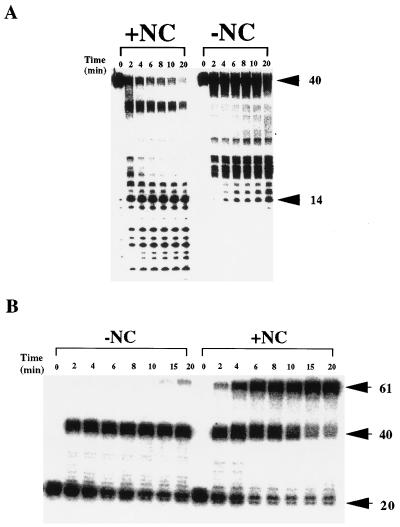
Time courses for strong-stop DNA synthesis and utilization in strand transfer by wild-type RT and p51 deletion derivatives are shown in Fig. 3B. The rate of accumulation of the 40-nt DNA product for each RT derivative was essentially the same as wild-type. However, the rate of utilization of the 40-nt DNA product in the strand transfer reaction was slightly diminished for both p66/p51Δ5 and p66/p51Δ9 and essentially abolished for p66/p51Δ13. The efficiency of strand transfer (the concentration of 61-nt DNA strand transfer product/concentration of 40-nt strong-stop DNA and 61-nt DNA strand transfer product) is changed relative to wild-type RT only for p66/p51Δ13. Based on the previous results, the defect in strand transfer for this enzyme is due to the inability of the synthesis-independent RNase H activity of this enzyme to hydrolyze the template RNA to oligomers of less than 14 nt. To test this possibility directly, a strand transfer reaction was completed with the Δ13 derivative by using a primary substrate containing a 5′ 32P-labeled 40-nt RNA. The results from this experiment are shown in Fig. 4A (panel −NC). As expected, the template RNA is rarely hydrolyzed to a product less than 13–14 nt.
Figure 4.
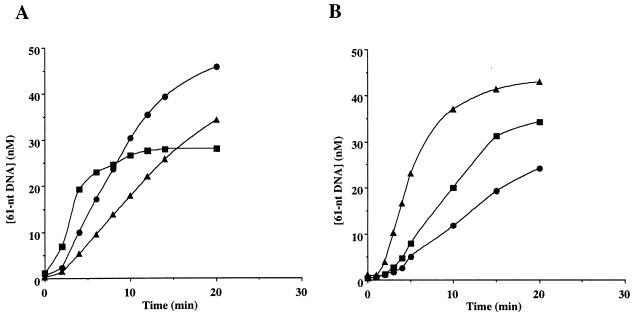
Effect of HIV NC on RNA hydrolysis and DNA strand transfer catalyzed by the p66/p51Δ13 RT. (A) Strand transfer reactions were completed as described with radiolabeled 40-nt RNA primary template (see Fig. 3A). RNase H hydrolysis products were resolved by electrophoresis on a denaturing 15% polyacrylamide gel. Reactions were completed in the presence (+NC) or absence (−NC) of the 71-amino acid form of HIV NC (10 μM). (B) Strand transfer reactions were performed as described with radiolabeled 20-nt DNA primer (see Fig. 3A). Quenched reactions were subjected to electrophoresis on a denaturing 15% polyacrylamide gel. Reactions were completed in the absence (−NC) or presence (+NC) of the 71-amino acid form of HIV NC (10 μM).
HIV NC Suppresses the Defects in the Synthesis-Independent RNase H Activity and Strand Transfer Observed for the Δ13 Mutant.
Peliska et al. (8) have shown that HIV NC stimulates the slow, synthesis-independent RNase H activity of HIV RT. While the basis for this stimulation is not known, these investigators speculated that a specific complex might exist between RT and NC. As a result, a marked increased in the rate of RT-catalyzed DNA strand transfer is observed in the presence of NC. Because the primary defect of p66/p51Δ13 is related to the synthesis-independent RNase H activity, we hypothesized that NC might modulate the RNase H activity of this enzyme. To test this hypothesis, a strand transfer assay was performed in the presence of concentrations of NC sufficient to completely coat all the RNA. NC was, indeed, capable of completely suppressing the defects observed with p66/p51Δ13 RT. As shown in Fig. 4A (+NC), in the presence of NC, template RNA was hydrolyzed to oligomers less than 14 nt (see Fig. 4A, −NC). Furthermore, as shown in Fig. 4B (+NC), NC reversed the defect in strand transfer observed with this RT derivative (see Fig. 4B, −NC). Quantitation of the strand transfer results demonstrated that the rate of strand transfer in the presence of NC for p66/p51Δ13 RT was identical to that of wild-type enzyme (data not shown). Finally, oxidized NC and the 55-amino acid form of HIV NC failed to suppress defects observed with the Δ13 mutant or to stimulate wild-type enzyme, regardless of the concentration used (data not shown).
EIAV NC Fails to Suppress Defects in p66/p51Δ13 RT Catalyzed DNA Strand Transfer.
The observation that the 55-amino acid form of HIV NC failed to stimulate the synthesis-independent RNase H activity and therefore the strand transfer activity of wild-type HIV RT is consistent with a specific interaction of RT with the carboxyl-terminal residues of NC. Therefore, we reasoned that if a specific RT–NC complex activated synthesis-independent RNase H activity of HIV RT, then NCs from other retroviruses would be ineffective. To test this idea, a strand transfer experiment was performed using NC purified from infectious EIAV. As shown in Fig. 5A, EIAV NC was incapable of removing the lag observed in the strand transfer reaction and did not produce premature termination of the strand transfer reaction as would be expected if the synthesis-independent RNase H activity of HIV RT had been activated by EIAV NC (8). EIAV NC slightly enhanced the rate of strand transfer owing, most likely, to the nucleic acid-annealing properties of this protein, and this enhancement was dependent on the concentration of EIAV NC (Fig. 5B).
Figure 5.
Comparison of the effect of HIV NC and EIAV NC on RT-catalyzed DNA strand transfer. (A) Strand transfer reactions were performed as described using wild-type RT. The concentration of the 61-nt DNA strand transfer product is shown as a function of time for reactions completed in the absence of NC (▴), in the presence of 10 μM HIV NC (▪), and 10 μM EIAV NC (•). The plateau observed in the reaction with HIV NC is caused by activation of the synthesis-independent RNase H activity of HIV RT which depletes the system of 41-nt acceptor template (8). (B) Reactions were completed as described in A in the presence of 0.5 μM (•), 2 μM (▪), and 8 μM (▴) EIAV NC.
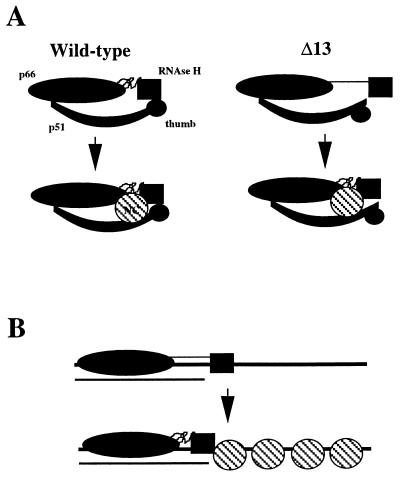
Taken together, these results are consistent with the model illustrated in Fig. 6A in which a specific interaction exists between the carboxyl-terminal amino acids of HIV NC and possibly the RNase H domain and p51 thumb of the HIV RT heterodimer. Implicit in this model is the requirement for NC to have a defined structure for formation of the complex with RT because oxidized NC fails to suppress the Δ13 defect. The existence of an RT–NC complex is also supported by genetic studies which have implicated NC in reverse transcription (16). It is worth noting that the strength of this interaction need not be significant because the effective concentration of NC in virions is in the millimolar range. In fact, if the interaction between RT and NC were too strong, DNA synthesis would probably be inhibited.
Figure 6.
Models for the suppression of the p66/p51Δ13 defect by HIV NC. (A) Specific Interaction between RT and NC. (Wild-type) The RNase H domain of the wild-type enzyme is stabilized by interactions with the p51 thumb, and this interaction is strengthened by the presence of NC which may interact with p66, the RNase H domain and p51. (Δ13) Deletion of the carboxyl-terminal residues of p51 alters the conformation of the p51 thumb, thus increasing the spatial relationship between the polymerase and RNase H catalytic centers. The wild-type conformation is induced into the p66/p51Δ13 RT in the presence of NC. (B) Nonspecific interaction between RT and NC. For clarity only the large subunit of the p66/p51Δ13 heterodimer is indicated. The nucleic acid that is illustrated represents strong-stop DNA (thick line) with residual template RNA (thin line) bound to the 3′ end of the DNA. The binding of p66/p51Δ13 RT to nucleic acid is dictated by the polymerase domain. The availability of nucleic acid for binding to the RNase H domain 26–27 nt from the polymerase active site permits stable binding of this enzyme to nucleic acid. In the presence of NC, the nucleic acid-binding domain of RNase H is restricted to interaction with the duplex region. In this case, the wild-type conformation is induced solely by the interaction of the RNase H domain with nucleic acid.
We have interpreted the lack of suppression activity of the 55-amino acid form of HIV NC to indicate that the carboxyl-terminal residues of NC mediates the interaction with RT. However, the biochemical properties of NC55 are different than that of the 71-amino acid form (17). For example, NC55 oxidizes more readily than NC (David Giedroc, personal communication). Therefore, oxidation is a trivial explanation for this result. It is worth noting that while NC55 is thought to be the major form of the NC protein found in virions, less than 10% of virion-associated NC protein would have to be of the 71-amino acid form to interact stoichiometrically with virion-associated RT. It is plausible that an interaction between RT and NC71 would preclude proteolytic processing of complexed NC71 in a manner analogous to that observed with the RT heterodimer.
An alternative model for the observed suppression of the defect in p66/p51Δ13 RT by NC, and stimulation of the synthesis-independent RNase H activity of wild-type RT, is based solely on the single-stranded, nucleic acid-binding activity of NC (Fig. 6B). In this model, the single-stranded portion of the strand transfer intermediate (i.e., strong-stop DNA hybridized to residual template RNA) is coated by NC and this restricts accessibility of the RNase H domain to the duplex region of the strand transfer intermediate (which is most likely free of NC) (16). This model seems unlikely, however, because EIAV NC, which displays nucleic acid-binding properties similar to that of HIV NC (18), cannot substitute for HIV NC (Fig. 5A).
While the changes observed with p66/p51Δ13 RT are quite pronounced, it is not clear whether they are caused by loss of a limited number of specific amino acid side-chain interactions or if all 13 amino acids must be removed to observe the effects described herein. It is clear, however, that deletion of all 13 amino acids is necessary to observe changes in the spatial relationship between the two catalytic centers of RT because enzymes in which 10, 11, or even 12 amino acids were removed from the p51 carboxyl terminus behave like wild-type RT (K. J. Howard and S.F.J.L.G., unpublished observations) Moreover, whether the change in the site of the first endonucleolytic cleavage of the RNA is due to a change in the position of the RNase H domain relative to the polymerase domain or due to repositioning of the nucleic acid in the enzyme cannot be determined unambiguously from the studies presented here. Attempts were made to use DNase I footprinting on DNA primer/templates to resolve this issue. While these studies demonstrated conclusively that the polymerase active site of p66/p51Δ13 was positioned properly over the primer 3′ hydroxyl, the precise location of the RNase H domain remained obscure (data not shown).
A role for the p51 subunit in primer/template and tRNA binding has been well documented (19–22). Also, Arts et al. (11) have shown that p66/p51Δ13 RT is impaired in its ability to disrupt loop–loop interactions between the lysyl-3 tRNA and HIV genomic RNA, presumably a function of the p51 subunit. Therefore, it is also possible to explain these results solely on the basis of changes that alter interactions of p51 with nucleic acid. This possibility does not seem likely because the magnitude of the change required to expand RNase H site selection, as observed here, should alter the stability of the RT–nucleic acid complex significantly, which was not observed (Table 1).
The fact that p66/p51Δ13 RT retains the ability to remove an additional 9 bases of RNA after the initial endonucleolytic cleavage in a manner identical to wild-type RT suggests that this activity is dictated solely by the RNase H domain. In support of this conclusion is the finding that only mutations encoding the RNase H domain itself have been shown to modulate this activity (13, 23). Conversely, in each case where this “directional-processing” activity of RT has been altered, no change in the site of the first endonucleolytic cleavage has been observed (13).
In conclusion, these studies ascribe a significant role to the carboxyl terminus of the p51 subunit of the HIV RT heterodimer—namely, that of a scaffold for the correct positioning of the RNase H active site relative to the polymerase active site. More importantly, the observation that HIV NC suppresses a defect associated with a deletion in the carboxyl terminus of p51 provides compelling evidence for the existence of a complex between these HIV proteins. This work and others (8, 24, 25) have shown that the primary role of the RT–NC complex is to activate the synthesis-independent RNase H activity thereby greatly diminishing the lag observed during RT-catalyzed DNA strand transfer in vitro. Evidence for such a lag in vivo does not exist. In fact, strong-stop DNA is never found inside infected cells (5). Finally, RNase H and strand transfer assays using the p51Δ13 derivative described here should prove useful in defining further elements of RT and NC necessary for formation of this complex.
Acknowledgments
We thank D. Giedroc for HIV NC, R. Gorelick for HIV NC55, and R. Montelaro for EIAV NC. C.E.C. is grateful to J. Arnold for assistance in preparation of the manuscript, D. Rodgers for preparation of Fig. 1B, and A. M. Skalka, D. Rodgers, A. Berdis, D. Sexton, and D. Smithrud for critical reading of the manuscript. This research was supported in part by National Institute of Health Grants AI09076 (to C.E.C.), GM13306 (to S.J.B.) and GM52263 (to S.F.J.L.G.).
ABBREVIATIONS
- RT
reverse transcriptase
- NC
nucleocapsid
- EIAV
equine infectious anemia virus
References
- 1.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauget C, Axler-Blin C, Vezinet-Brun F, Rouzious C, Rozenbaum W, Montagnier L. Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Popovic M, Sarngadharan M G, Read E, Gallo R C. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 3.Coffin J M. In: Virology. Fields B N, editor. New York: Raven; 1990. pp. 1437–1500. [Google Scholar]
- 4.Larder B A. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 205–222. [Google Scholar]
- 5.Telesnitsky A, Goff S P. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 49–83. [Google Scholar]
- 6.Peliska J A, Benkovic S J. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 7.Gopalakrishnan V, Peliska J A, Benkovic S J. Proc Natl Acad Sci USA. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 9.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacques P S, Woehrl B M, Howard K J, Le Grice S F J. J Biol Chem. 1994;269:1388–1393. [PubMed] [Google Scholar]
- 11.Arts E J, Ghosh M, Jacques P S, Ehresmann B, Le Grice S F J. J Biol Chem. 1996;271:9054–9061. doi: 10.1074/jbc.271.15.9054. [DOI] [PubMed] [Google Scholar]
- 12.Kati W M, Johnson K A, Jerva L F, Anderson K S. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 13.Ghosh M, Howard K J, Cameron C E, Benkovic S J, Hughes S H, Le Grice S F J. J Biol Chem. 1995;270:7068–7076. doi: 10.1074/jbc.270.13.7068. [DOI] [PubMed] [Google Scholar]
- 14.Metzger W, Hermann T, Schatz O, Le Grice S F J, Heumann H. Proc Natl Acad Sci USA. 1993;90:5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woehrl B M, Georgiadis M, Telesnitsky A, Hendrickson W, Le Grice S F J. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]
- 16.Darlix J-L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 17.Khan R, Giedroc D P. J Biol Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- 18.Gelfand C A, Wang Q, Randall S, Jentoft J E. J Biol Chem. 1993;268:18450–18456. [PubMed] [Google Scholar]
- 19.Arts E J, Stetor S R, Li X, Rausch J W, Howard K J, Ehresmann B, North T W, Woehrl B M, Goody R S, Wainberg M A, Le Grice S F J. Proc Natl Acad Sci USA. 1996;93:10063–10068. doi: 10.1073/pnas.93.19.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermann T, Meier T, Goette M, Heumann H. Nucleic Acids Res. 1994;22:4625–4633. doi: 10.1093/nar/22.22.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohlstaedt L A, Wang J, Rice P A, Friedman J M, Steitz T A. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 223–249. [Google Scholar]
- 22.Mishima V, Steitz J. EMBO J. 1995;14:2679–2687. doi: 10.1002/j.1460-2075.1995.tb07266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausch J W, Arts E J, Woehrl B M, Le Grice S F J. J Mol Biol. 1996;257:500–511. doi: 10.1006/jmbi.1996.0181. [DOI] [PubMed] [Google Scholar]
- 24.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You J C, McHenry C S. J Biol Chem. 1994;269:31491–31495. [PubMed] [Google Scholar]