Abstract
Escherichia coli cells lacking the histone-like protein HU form filaments and have an abnormal number of anucleate cells. Furthermore, their phenotype resembles that of rfa mutants, the well-characterized deep-rough phenotype, as they show an enhanced permeability that renders them hypersensitive to chloramphenicol, novobiocin, and detergents. We show that, unlike rfa mutants, hupAB mutants do not have a truncated lipopolysaccharide but do have an abnormal abundance of OmpF porin in their outer membrane. While the complete absence of HU does not abolish the osmoregulation of OmpF protein synthesis, the steady-state level of micF RNA, the negative regulator of OmpF, decreases in bacteria lacking HU, increasing the basal level of this membrane protein. These findings demonstrate a novel link between a bacterial chromosomal protein and the outer membrane composition.
Keywords: bacterial chromosomal protein, hypersensitivity to antibiotics, OmpF porin, micF RNA
The Escherichia coli HU protein is one of the most abundant DNA-binding proteins associated with the bacterial chromosome (1, 2). This small, basic, dimeric protein, composed of two closely related subunits, shares with histones the ability to introduce negative supercoiling into relaxed DNA molecules in the presence of topoisomerase I in vitro (3). This property makes HU one of the best candidates for constraining DNA supercoils in the bacterial chromosome. In addition, it has recently been shown that a relationship exists between the level of HU protein in vivo and the activity of DNA topoisomerase I. Thus, in addition to restraining negative supercoils, HU may, in vivo, communicate with topoisomerase I to regulate the global level of supercoiling in E. coli (4). Furthermore, biochemical and genetic studies have shown that HU participates in several specific processes, including oriC-dependent DNA replication, hin-mediated gene inversion, transposition of bacteriophage Mu, and the transposon 10 and DNA repair (5–9), probably as a component of active multiprotein complexes.
The hupB gene encoding the HUβ subunit and the hupA gene encoding HUα subunit were isolated and, respectively, mapped at 10.5 and 90.7 min on the E. coli chromosome (10, 11). To better understand the role of HU in vivo we attempted to construct mutants lacking this protein (12). The hupB and the hupA genes were, respectively, disrupted by a kanamycin and a chloramphenicol-resistance cassette. Surprisingly, E. coli cells survived, albeit poorly, in the absence of HU. These mutants, as well as those constructed by Wada et al. (13), exhibited perturbations of growth and cell division and defects in transposition of phage Mu. Hence, the evidence seemed to show that HU was not essential for growth. In the course of this work, we observed an astonishing property of these double mutants. When plated on Luria–Bertani (LB) medium agar containing chloramphenicol, the hupAB mutants exhibited extreme sensitivity to chloramphenicol. This sensitivity was not observed with the single hupA mutant even though both contained the same chloramphenicol-resistance cassette. To check if this sensitivity was due to a low expression of chloramphenicol acetyltransferase carried by the resistance cassette, we measured the level of chloramphenicol acetyltransferase activity in sonicated extracts and found no difference between the hupA and hupAB mutants (12). Therefore, we concluded that the permeability to chloramphenicol was much increased in the hupAB mutant compared with hupA strains. In the course of further work using these double mutants, several striking facts highlighted the fragility of the double hupAB mutants. For these reasons, we investigated a possible change in the cell envelope composition of the hup mutants.
The envelope of Gram-negative bacteria consists of three layers: the outer membrane (OM), the peptidoglycan, and the inner cytoplasmic membrane. The inner membrane contains most of the transport systems and machinery for protein export. The peptidoglycan is a large heteropolymer that confers the rigidity of the cell envelope, hence the shape of the cell, and protects it from osmotic lysis. In Gram-negative bacteria, it consists of a network of amino sugars and amino acids. The OM, which plays an important role in the physiology of these bacteria, is a strong permeability barrier for all nutrients (and/or antibiotics) on their way from the medium to the periplasm and prevents leakage of periplasmic proteins. In enterobacteria, the OM is an asymmetric bilayer located outside the peptidoglycan. It is composed of glycerophospholipids, lipopolysaccharides (LPS), and proteins that are responsible for selective permeability to nutrients and antibiotics. This function is fulfilled by a few major proteins called porins.
We show here that the increased sensitivity to antibiotics and detergents of the hupAB mutants is related to an overexpression of OmpF, one OM porin. This high level of OmpF is due to a decreased accumulation of micF RNA, a negative regulator of OmpF translation.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Growth Conditions.
The bacteria used in this study were E. coli K12, C600 (F- thr leu tonA rpsL supE lacY) and its hup derivatives, which were described by Huisman et al. (12). Unless otherwise noted, cells were grown in LB medium (10 g of tryptone/5 g of yeast extract/5 g of NaCl per liter) at 37°C. “LB-0” corresponds to the LB medium without NaCl. The ompC and ompF mutants were, respectively, MH225 (MC4100 malQ7 Φ(ompC::lacZ) 10–25 and MH 513 (MC4100 araD+ Φ(ompF::lacZ) 16–13. The plasmid pompF is a kanamycin-resistant tetracycline-sensitive derivative of pLG361 (14). The BamHI–BamHI fragment from pUC4K (carrying the kanamycin cassette) was inserted into the BamHI site of pLG361.
Osmolarity-Dependent Tetracycline Sensitivity.
Exponential cultures of strains C600 and C600 hupAB were diluted 100-fold in “liquid” LB agar medium without (0 mM) or with (300 mM) NaCl and quickly poured into Petri dishes. Whatman 3MM 5-mm disks were soaked into tetracycline solution (0.1 mg/ml in 50% ethanol) and then deposited on top of the solidified agar. Plates were incubated overnight at 37°C. The halo diameter represents the relative sensitivity to the antibiotic.
Temperature-Dependent Tetracycline Sensitivity.
Five microliters of strains C600, C600 hupAB, and C600 transformed by pompF from overnight cultures in LB medium were spotted on LB plates with or without tetracycline (1 μg/ml) and incubated overnight at 30°C, 37°C, and 42°C.
Phenotype Tests.
Exponential cultures of C600 (wt), hupB, hupA, and hupAB were diluted (1:100) in LB liquid medium with an increasing concentration of cholic acid (3α,7α,12α-trihydroxy-5b-cholan-24-oic acid; Sigma) (0–15%) or SDS (0–3%). Viability was assayed by measuring the absorbance at 600 nm after overnight incubation.
Extraction of the OM Proteins (OMP).
C600 and C600 hupAB were grown overnight in LB liquid medium at 37°C. Then an aliquot corresponding to 50 OD600 was centrifuged. The pellets were washed twice in 10 ml of TH buffer (10 mM EDTA/Hepes/10 mM NaOH, pH 7.4). The cells were then disrupted using a French press (30,000 psi) three times. Cellular debris was removed by centrifugation for 10 min at 10,000 × g and the supernatant (10 ml) was collected. The inner membrane was solubilized by incubation of the lysate with Triton X-100 (1%) and MgSO4 (1 mM) for 30 min on ice as described by Pugsley and Schnaitman (15). The Triton X-100 insoluble fraction containing the OM was recovered by centrifugation for 60 min at 15,000 rpm. The membranes were resuspended in 200 μl of TH buffer and 5 μg were mixed with an equal volume of sample buffer 2 times, boiled for 5 min at 100°C, and analyzed on 8% SDS polyacrylamide gels containing 8M urea. Migration was for 6 hr at 150 V at room temperature.
Analysis of mRNAs.
Total RNA was extracted from cells (10 OD600 units total) and grown overnight in LB or LB-0 medium by the hot-phenol procedure (16). Extension of cDNA was done using synthetic oligonucleotides as primers. They were for ompF–RT, 5′-GACACCTGCCACTGCCG; ompC–RT, 5-′GCAGGCCGTCTAGTTTACCG; and micF–RT: 5′-GGGGTAAACAGACATTCAG. The 5′ end of the oligonucleotide was labeled with [gamma-32P]-ATP (3,000 Ci/mmol; 1 Ci = 37 Gbq) and T4 polynucleotide kinase (Biolabs, Northbrook, IL). Total RNA (15 μg) was annealed with 10 ng of labeled primer for 5 min at 70°C in H2O plus an additional 5 min in RTB 1× (50 mM Tris⋅HCl, pH 8.3/40 mM KCl/6 mM MgCl2) and then cooled slowly at room temperature. Synthesis of cDNA was performed in the same buffer for 30 min at 45°C in the presence of nonradioactive deoxynucleotides at a concentration of 225 μM each and 1 unit of reverse transcriptase RAV-2 (Amersham). The reaction was then stopped by the addition of 100 μl of 3 M sodium acetate and 250 μl of ethanol, precipitated at −20°C, centrifugated, and resuspended in formamide dye solution. The products were analyzed on a 8% polyacrylamide/8 M urea sequencing gel and then autoradiographed. To obtain quantitative results, reverse transcriptase and all substrates except the template RNA were used in excess. The total amount of RNA used was visualized after migration on agarose gels and ethidium bromide staining.
RESULTS
HU Mutants Resemble the Deep-Rough Phenotype.
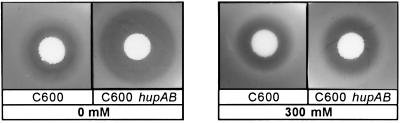
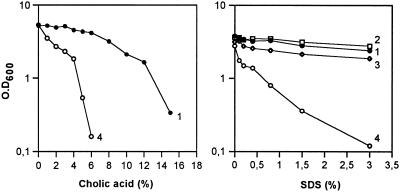
During the construction of the hup mutants we found that the double hupAB mutant lacking the two subunits of HU was abnormally sensitive to chloramphenicol, even though it contained the resistance gene. In fact, a difference of nearly two logs in viability on LB/agar plates, containing chloramphenicol (12.5 μg/ml) and NaCl (5 g/liter), was found between the hupAB and hupA mutants, although both had their hupA gene interrupted with a chloramphenicol-resistance cassette. This sensitivity was not due to a defect in the synthesis of the acetylase, which confers resistance to this antibiotic (12). A further observation was that hupAB mutants were not only hypersensitive to chloramphenicol, even when the cassette was perfectly expressed, but were generally oversensitive to all the antibiotics tested, even when present at a very low concentration. This suggested that the double mutant may have increased permeability. It is possible that defects in the cell envelope could have rendered these mutants more fragile or more sensitive to drugs. It was intriguing that this hypersensitivity was almost eliminated when the NaCl concentration of the LB agar plates was increased from 5 g/liter to 10 g/liter, suggesting that salt could somehow compensate for this fragility in the hupAB mutant. Fig. 1 illustrates the hypersensitivity of hupAB cells to a low concentration of tetracycline (1μg/ml) compared with wild-type cells in low salt medium. In the presence of 300 mM NaCl, the observed difference due to the absence of HU is virtually undetectable. Similarly, the hupAB mutant, constructed in a C600 background, grew very poorly when plated on MacConkey–lac agar plates containing a low amount of NaCl, compared with hup+ cells or single hup mutants, reinforcing the possibility of real changes in the cell envelope of these mutants (data not shown). To further examine this possibility, sensitivity to bile salts and detergents such as cholic acid and SDS, was measured. Fig. 2 illustrates the dramatic sensitivity of hupAB mutants to cholic acid and to SDS in LB liquid medium, whereas the growth of hup+, hupB, and hupA cells was unaffected by 3% SDS.
Figure 1.
Hypersensitivity of the hupAB strain to tetracycline at low osmolarity. The halo diameter represents the relative sensitivities of C600 and C600 hupAB to the antibiotic tetracycline (1μg/ml) contained in Petri dishes. The NaCl concentration of the medium, 0 mM or 300 mM, represents the osmolarity of the solid medium.
Figure 2.
Hypersensitivity of hupAB strain to detergent and bile salts. C600 and its hup derivatives were grown in LB supplemented with cholic acid or SDS. Samples were: (1) C600 (wt); (2) C600 hupB; (3) C600 hupA; (4) C600 hupAB.
Sensitivity to antibiotics, detergents, and cold (J.R.-Y., unpublished data) strongly suggested that the hupAB mutants might have a “deep-rough phenotype.” This phenotype characterizes some rfa deletion mutants of E. coli, which have mutations in genes implicated in the synthesis of LPS (17). These mutants, which synthesize a truncated LPS core, have modified cell surface properties. Similar to the hupAB mutants, they are hypersensitive to tetracycline, chloramphenicol, novobiocin, and detergents. Furthermore, like the deep-rough mutants, the hupAB cells form filaments (12). The two phenotypes thus being very similar, we analyzed the LPS of these mutants. The OM of E. coli is asymmetric because the LPS is normally found only in the outer leaflet and interacts with the environment, whereas the inner leaflet is composed of phospholipids. The lipid portion (lipid A) of the LPS has been shown to be responsible for most of the biological activities of these macromolecules, whereas total loss of the O-polysaccharide part affects the permeability of the membrane (18). The deep-rough mutants of E. coli have lost most, or all, core sugars except ketodeoxyoctulosonate.
The LPS fractions, isolated from both C600 and C600 hupAB cells, were qualitatively compared with each other and to a standard (E. coli Ra strain) by three different methods. Thin layer chromatography (19) gave unsatisfactory resolution of the samples, but similar Rfs (relative electrophoresis mobility). Analysis by SDS/PAGE showed unambiguously that the hupAB LPS were not of the deep-rough (Re) mutant type. The LPS of the three strains all migrated roughly to the same level. If some heterogeneity was observed it was observed in all three LPS and may be due to small variations between the standard strain and C600, but not due to modification of the core sugars. Finally, mass spectrometry (20) of both the isolated lipids and polysaccharides moities of LPS indicated that each lipid corresponded to the classical E. coli type hesaacyl lipid A and that the polysaccharide cores were normal. These experiments (data not shown) demonstrated that the hupAB mutants are not deep-rough (Re) mutants but full-core (Ra) strains.
OmpF Is Overproduced in hupAB Mutants at Low Ionic Strength.
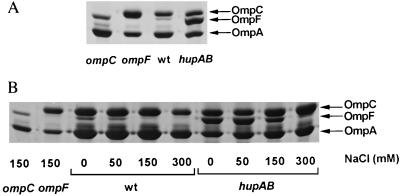
Because the extreme permeability observed in the cells lacking HU could not be explained by change in the LPS, we analyzed the membrane proteins of the hupAB mutants. First, we studied the proteins extracted from total membranes. The comparison between the wild-type and the hupAB mutant showed several differences in the electrophoretic profiles on SDS/10–25% PAGE in the range where the porins, produced in very large amounts, migrate (data not shown). Consequently, we specifically isolated and analyzed the OMP. Extraction of these porins was carried out using their property of insolubility as described by Pugsley and Schnaitman (15). Fig. 3A shows the marked difference in the profiles of the major porins extracted from a hupAB mutant as compared with the wild-type strain. To unambiguously identify the major porins, OmpC and OmpF on these gels, we performed parallel analysis of extracts prepared from the ompC and ompF mutants prepared under the same conditions. Clearly, there is a major change in the OmpF to OmpC ratio. In the wild-type strain, C600, a low level of OmpF is present compared with high levels of OmpC and OmpA, whereas in the absence of HU, under the same conditions of growth, OmpF is clearly the most abundant porin present in the OM. Immunoblotting with a serum prepared against the OmpF protein proved the identification of these two major OMPs extracted from the hupAB mutant (data not shown). In this figure, the amount of OmpC seems slightly decreased in the hupAB mutant, but certainly not as much as would be expected if the total amount of OmpC and OmpF together was constant (21). Several experiments showed that OmpC is not very strongly affected by the absence of HU (data not shown). Other abundant porins such as OmpA did not seem to be affected at all.
Figure 3.
Profile of porins extracted from hup+ (wt) or hupAB strains grown at different ionic strength. (A) OMPs were extracted from different strains grown in LB medium and analyzed by gel electrophoresis as described. Samples were MH225 (ompC), MH513 (ompF), C600 (wt), and C600 hupAB. (B) Extracts of porins from strains C600 (wt) and hupAB mutant grown in LB at the different ionic strengths indicated were analyzed on a 12% acrylamide gel with 7 M urea. MH225 (ompC), MH513 (ompF), C600 (wt), C600 hupAB.
Since the total amount of OmpF increased sharply in the absence of HU, we wondered whether the basal level of OmpF protein was dependent on the total amount of HU protein in the cell. Using wild-type hup+, hupB, hupA, or hupAB strains, we obtained cells with, respectively, 100, 50, 10, or 0% of HU (the HUβ subunit is partially unstable in the absence of the α subunit) (22). Extraction of the OMPs from these strains and their analysis by PAGE showed that the effect on OmpF synthesis was observed predominantly in the hupAB mutant (not shown).
How could the absence of HU modulate the level of the OMPs, specifically the expression of OmpF? The genes encoding for the abundant porins are under different regulatory mechanisms. The relative amounts of the OmpF and OmpC proteins are determined by many environmental factors including osmolarity, ionic strength, composition of the culture medium, the carbon source, pH, temperature, etc. (for review see ref. 23). We wondered at first if the absence of HU could alter the osmoregulation of the porins leading to a decreased expression of OmpF at high osmolarity, while OmpC levels stay constant or increase (24). To test this, we extracted porins from wild-type and hupAB bacteria that were grown in media with increasing NaCl concentrations. As can be seen in Fig. 3B, OmpF accumulation decreased with the increasing osmolarity of the media. In hupAB cells the great abundance of OmpF also decreased with osmolarity, but perhaps not as strongly as in the wild-type strain. Clearly the absence of HU, which sharply increased the level of OmpF, did not abolish its down-regulation at high-salt concentrations. This certainly explains why the hypersensitivity to tetracycline of the HU double mutant was observed only when the bacteria were plated on low-salt media (Fig. 1). The absence of HU had no effect on OmpC accumulation at high salt conditions. Similar results were obtained with sucrose, another osmolyte (data not shown).
Hypersensitivity of hupAB to Antibiotics Is Linked to Increased OmpF Level.
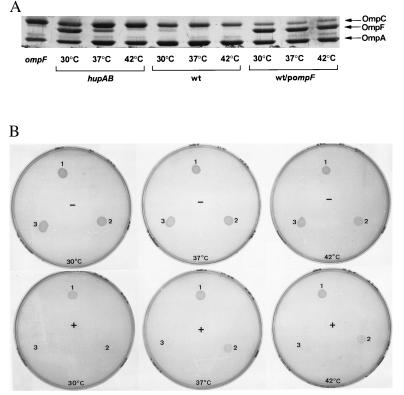
In addition to osmolarity, the temperature of growth strongly affects OmpF accumulation in the cell: the level of OmpF is higher at 30°C that at 37°C (25). To check if the OmpF overproduced in the hupAB mutant, due to the absence of HU, was also sensitive to the temperature of growth, we compared the profiles of porins extracted from either hup+ or hupAB cells grown at 30°C, 37°C, or 42°C. Fig. 4A shows that OmpF synthesis in the hupAB strain is also strongly temperature sensitive. The high level present in hupAB grown at 30°C is reduced at 37°C and disappears at 42°C. Interestingly, the OmpF overproduced by transforming a wild-type cell with a multicopy plasmid carrying the ompF gene is insensitive to temperature (Fig. 4A). This difference in temperature sensitivity clearly distinguishes the hupAB cells from the “wt/pompF” strain.
Figure 4.
Hypersensitivity to tetracycline and OmpF overproduction. (A) Five micrograms of total OMPs extracted from the strains C600 hupAB, C600, and C600 transformed by pompF grown in LB medium at 30°C, 37°C, and 42°C were analyzed by gel electrophoresis as described. (B) Five microliters of strains C600 (1), C600 hupAB (2), and C600 transformed by pompF (3) from an overnight culture in LB medium were spotted on LB plate with (+) or without (−) tetracycline (1 μg/ml) and incubated overnight at 30°C, 37°C, and 42°C.
OmpF and OmpC, the two major OMPs, function as passive diffusion pores for small, hydrophilic proteins into the periplasm. Although their structural and functional properties are similar they differ in pore size. We wondered if the hypersensitive phenotype to chloramphenicol and to tetracycline that we have described for the hupAB mutants could be associated with the overproduction of OmpF observed in these mutants. To test this hypothesis, we analyzed the effects produced by the hup+ strain transformed with a plasmid carrying the ompF gene. Fig. 4 shows that the overexpression of OmpF in these cells at all temperatures (Fig. 4A) makes the wild-type cell hypersensitive to antibiotics, in this case tetracycline. No difference in the degree of hypersensitivity is seen whatever the temperature of growth (Fig. 4B). The effects caused by the absence of HU have been shown to be particularly marked at low salt concentrations (on LB-0; Fig. 3B). Fig. 4 A and B show that the temperature of growth also affected this susceptibility to tetracycline. In the cells lacking HU the hypersensitivity to tetracycline declined with the same pattern than OmpF expression. The highest expression occurs at low temperature, decreases from 30°C to 37°C and disappears at 42°C. The temperature dependence of OmpF expression and hypersensitivity to tetracycline observed in the hupAB mutant, which was not observed when OmpF was present on a multicopy plasmid, makes the correlation between OmpF production and susceptibility to antibiotics very strong.
HU Acts on OmpF via micF.
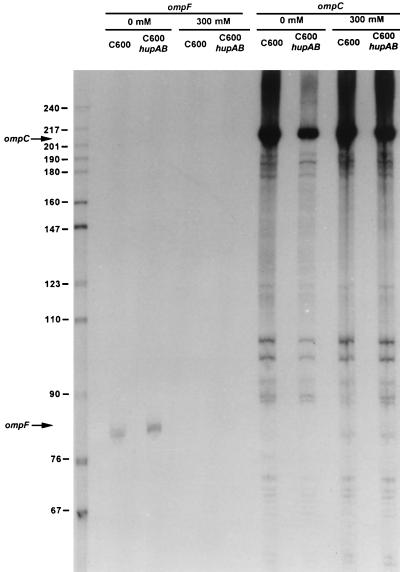
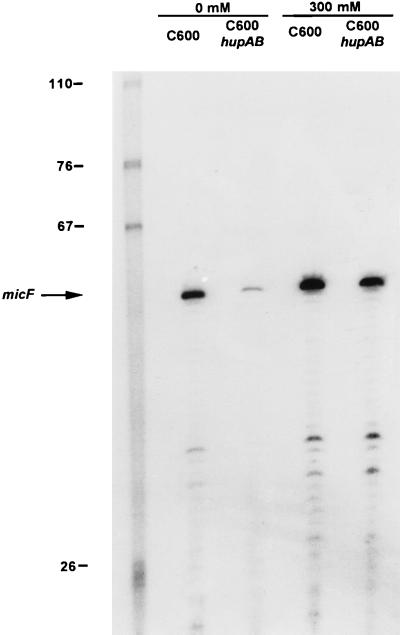
Osmoregulation of OmpF and OmpC expression, and probably regulation by other environmental factors, occurs at the transcriptional level and is controlled by a two component regulatory system, EnvZ (an osmosensory kinase) and OmpR (a positive regulator) (26). Another regulatory mechanism of porins has been described, which acts at the posttranscription level. The micF antisense RNA transcribed from a region located upstream from the ompC gene, but in the opposite direction to the ompC gene, forms an RNA–RNA hybrid with the 5′ end of the ompF transcript, thereby decreasing the rate of OmpF translation (27). To investigate whether the increased level of OmpF observed in the absence of HU involved transcriptional or translational control of ompF RNA we extracted the total mRNA from hup+ and hupAB strains grown either in the absence or in the presence of 300 mM NaCl. We analyzed first the level of ompF RNA from both wild-type and hupAB cells by a primer extension–reverse transcriptase-based assay using the ompF–RT oligonucleotide labeled at the 5′ end. Fig. 5 shows that in the absence of HU there is a small increase in the level of ompF RNA (1.5-fold). Similar experiment performed with ompC–RT oligonucleotide as the primer showed (Fig. 5) that symmetrically OmpC RNA decreased by a factor of two in the absence of NaCl. These results show that at the RNA level the regulation that directs an opposite variation in the OmpC–OmpF level, functions even in the absence of HU. These experiments also showed that osmoregulation switched off the transcription of ompF independently of HU, whereas ompC transcription was, also in both types of cells, at its highest level at 300 mM NaCl. However, by itself, these mild transcriptional effects could not explain the large increase in OmpF accumulation observed in the hupAB mutants (see Figs. 3 A and B, and 4A). To further investigate this issue, we measured the concentration of the micF RNA using the micF–RT oligonucleotide as a primer. When the bacteria were grown in LB with no salt, the level of micF RNA was strongly reduced (around 6-fold) in the absence of HU, whereas no difference was observed when the bacteria were grown in LB containing 300 mM NaCl (Fig. 6). This outcome clearly demonstrates that the absence of HU results in a decrease in the concentration of micF RNA in cells growing in low osmolarity medium and causes, as a consequence, an increase in OmpF synthesis. This effect is not observed when bacteria are grown at high osmolarity.
Figure 5.
Expression pattern of ompF and ompC mRNAs. C600 (wt) and C600 hupAB were grown in LB broth without NaCl (0) and with NaCl (300 mM). Total RNAs (15 μg) were subjected to primer extension analysis with the 5′ end-labeled oligonucleotide ompF–RT and ompC–RT. The RNA extension products were analyzed by electrophoresis on an 8% sequencing gel and then autoradiographed. Lane 1 represents the molecular weight markers and the other samples are as indicated.
Figure 6.
Expression pattern of micF mRNA. C600 (wt) and C600 hupAB were grown in LB broth without NaCl (0) and with NaCl (300 mM). Total RNAs (15 μg) were subjected to primer extension analysis with the 5′ end-labeled oligonucleotide micF–RT. The RNA extension products were analyzed by electrophoresis on an 8% sequencing gel and then autoradiographed. The first lane represents the molecular weight markers and the other samples are as indicated.
DISCUSSION
In the course of the construction of the double hupAB mutant, we observed that the cells lacking the protein HU were abnormally sensitive to antibiotics, bile salts, and detergents, particularly in growth medium of low osmolarity. The aim of this study was to investigate the causes of the membrane permeability defects observed in hupAB mutants and to understand at what level HU could act to affect the permeability barrier of E. coli.
The major function of the bacterial cell envelope, besides protecting the cell from the environment, is to permit and control the exchange and communication between the environment in which the bacteria live and the interior of the cell. In Gram-negative bacteria, several layers of the cell envelope contribute to preventing the free diffusion of hydrophobic solutes or compounds with a molecular weight greater than 600 daltons. One of these layers is the outside leaflet of the OM, which contains LPS molecules and different porins. Loss of part of the LPS structure, as seen in rfa mutants, leads to the well-known deep-rough phenotype, which confers an increase in sensitivity to detergents and hydrophobic antibiotics such as chloramphenicol and novobiocin. Many deep-rough mutants also exhibit a mucoid phenotype, which is caused by the production of a colanic acid capsular polysaccharide. We should recall that in a previous study it was shown that the deregulation of HU synthesis induces mucoidy (28). However, the mechanisms that cause the hypersensitivity of the deep-rough and of the hupAB mutants differ totally. Our results show that neither the biosynthesis of peptidoglycan implicated in cell wall formation (data not shown) nor the biosynthesis of the LPS were affected. The deep-rough mutant also leads to an elevation of the phosphatidylethanolamine content in the OM coupled with a reduction in porin proteins, particularly OmpF. In contrast, the hypersensitivity of the hupAB strains is shown here to be due to a strong increase in OmpF content.
The two major porins associated with peptidoglycan in the assemblage of the OM are OmpC and OmpF. The relative amount of the two proteins is dependent on the growth medium and on other environmental factors in such a way that the sum of their concentrations is almost constant. The two major regulatory pathways of these two porins are well established. The EnvZ–OmpR system regulates OmpC and OmpF at the transcriptional level (21). EnvZ, an inner membrane protein that has both a kinase and a phosphatase activity, senses the environment and transmits the signal from the medium to OmpR, a positive regulator that binds specific DNA sequences upstream from the ompF and ompC promoters. This system is thought to be implicated in the regulation of porins by osmolarity. OmpF is preferentially synthesized in media of low osmolarity and OmpC in media of high osmolarity (29). Our present data clearly show that the osmoregulation of OmpC and OmpF is not abolished by the absence of HU. Another mechanism may be superimposed on the regulation by EnvZ–OmpR. Increase in osmolarity seems to increase DNA negative supercoiling (30). Mutations that alter DNA supercoiling show that porin expression is in fact sensitive to the level of DNA supercoiling, but a perfect correlation between supercoiling changes and porin expression was not observed (24). Interestingly, an increase in OmpC and a decrease in OmpF level in the absence of OsmZ (H-NS) was observed (23). HU, which shares with H-NS the property of introducing, in vitro, negative supercoiling in relaxed DNA in the presence of topoisomerase I (3, 31) has clearly opposite effects on OmpF. Moreover, the effect of HU could not occur through a change in DNA supercoiling, since it was shown that in E. coli the absence of HU causes only a marginal change in the superhelical density of the DNA: the absence of HU being correlated with an increased activity of topoisomerase I (4).
The second regulatory circuit affecting the expression of OmpC–OmpF operates at the translational level and involves micF, a small antisense RNA (27). This small RNA produced by an independent transcriptional unit located upstream from ompC, but transcribed in the opposite direction, is complementary to the 5′ portion of ompF mRNA and therefore inhibits its translation. We show here that the amount of micF is significantly reduced in the hupAB mutant at low salt. This certainly explains the increase observed in OmpF level. Here again, the absence of HU has a relatively small effect on the synthesis of micF RNA at high salt growth medium. We do not yet know whether it is by a direct or indirect mechanism that HU affects micF. It was shown that the binding of IHF, the integration host factor (32), to a region upstream from OmpC has a negative effect on in vitro transcription of OmpC (33). More recently, the OmpF level was found to be very high in IHF mutants but in contrast to HU, the absence of IHF, by affecting the transcriptional activity of ompF, alters the osmoregulation of OmpF. IHF is required for the decrease in OmpF expression observed at high osmolarity, HU is not (34, 35).
Probably most intriguing aspect is the resemblance between the properties of bacteria mutated in the mar locus and those of cells lacking HU, such as the decrease observed in the level of the small micF RNA, which leads to the increase in the amount of OmpF and increased susceptibility to antibiotics. In enterobacteria, the mar locus has been found to mediate intrinsic multiple antibiotic resistance (36). The mar mutants or mutants that overexpress marA, one of the genes of this locus, are more resistant to antibiotics and are insensitive to the killing effects of fluroquinolones (37). On the other hand, deletion of this locus or inactivation of the marA gene (marA::Tn5) (38) leads to increased susceptibility to multiple antibiotics. Interestingly, like the “HU-phenotype,” the mar phenotypes are linked to changes in both OmpF protein and micF RNA. Resistance to antibiotics is correlated with a decrease in the OmpF level, whereas the OmpF level increases in susceptible strains. Moreover, it has been shown that the decreased level of OmpF in mar mutants is due to an increased expression of micF. The hypersensitive phenotype that we have described for the hupAB mutants corresponds to that described for strains that lack the marA locus. The fact that the hupAB mutants that are hypersensitive to chloramphenicol and novobiocin are also, like the mar deletion mutants, hypersensitive to tetracycline certainly supports this hypothesis. Again, these experiments clearly demonstrate the strong correlation between the abnormal susceptibility of E. coli cells to antibiotics and the abnormal presence of OmpF in the bacteria. Further studies are essential to clarify if HU and the marA gene product regulate micF by a similar mechanism and what the interrelationship between these two systems is. Future experiments should clarify whether the effect of HU on micF synthesis is direct or mediated via the mar locus.
The conditions prevailing in the digestive tract, the natural environment of E. coli (high osmotic pressure, high temperature, low pH), favor the synthesis of OmpC, a more desirable porin in such an environment because it produces a slightly smaller channel. Our results show that HU participates in a control pathway that down-modulates OmpF. In its absence, enterobacteria like E. coli will fail to grow in its natural host.
Acknowledgments
We are greatly indebted to D. Mengin-Lecreulx for advice and helping us with peptidoglycan analysis. We are grateful to C. Wandersman, T. Pugsley, T. Silhavy, and D. Mengin-Lecreulx for gifts of serum, strains, and/or numerous helpful discussions and constructive criticism of this manuscript. The plasmid pLG361 was a gift from C. Lazdunski and R. Lloubes. This work was supported by the Centre National de la Recherche Scientifique and grants from l’Association de la Recherche contre le Cancer (ARC) and Commission of the European Communities (CEC) (HCM project CHRX-CT92–0010). E.P. was a recipient of a Ph.D. fellowship from the Ministére de la Recherche et de l’Espace (Paris VI), ARC, and Les Amis des Sciences.
ABBREVIATIONS
- OM
outer membrane
- OMP
outer membrane protein
- LPS
lipopolysaccharide
- IHF
integration host factor
References
- 1.Rouviere-Yaniv J, Gros F. Proc Natl Acad Sci USA. 1975;72:3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouviere-Yaniv J. Cold Spring Harbor Symp Quant Biol. 1978;42:4435–4450. doi: 10.1101/sqb.1978.042.01.047. [DOI] [PubMed] [Google Scholar]
- 3.Rouviere-Yaniv J, Yaniv M, Germond J E. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 4.Bensaid A, Almeida A, Drlica K, Rouviere-Yaniv J. J Mol Biol. 1996;256:292–300. doi: 10.1006/jmbi.1996.0086. [DOI] [PubMed] [Google Scholar]
- 5.Dixon N E, Kornberg A. Proc Natl Acad Sci USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson R C, Bruist M F, Simon M I. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 7.Craigie R, Arndt-Jovin D J, Mizuuchi K. Proc Natl Acad Sci USA. 1985;82:7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morisato D, Kleckner N. Cell. 1987;51:101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 9.Boubrik F, Rouviere-Yaniv J. Proc Natl Acad Sci USA. 1995;92:3958–3962. doi: 10.1073/pnas.92.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kano Y, Wada M, Nagaso T, Imamoto F. Gene. 1986;45:37–44. doi: 10.1016/0378-1119(86)90129-0. [DOI] [PubMed] [Google Scholar]
- 11.Kano Y, Osato K, Wada M, Imamoto F. Mol Gen Genet. 1987;209:408–410. doi: 10.1007/BF00329674. [DOI] [PubMed] [Google Scholar]
- 12.Huisman O, Faelen M, Girard D, Jaffe A, Toussaint A, Rouviere-Yaniv J. J Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada M, Kano Y, Ogawa T, Okasaki T, Imamoto F. J Mol Biol. 1988;204:581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- 14.Jackson M E, Pratt J M, Stoker N G, Holland I B. EMBO J. 1985;4:2377–2383. doi: 10.1002/j.1460-2075.1985.tb03942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugsley A P, Schnaitman C A. Biochim Biophys Acta. 1979;581:163–178. doi: 10.1016/0005-2795(79)90233-2. [DOI] [PubMed] [Google Scholar]
- 16.Qu H L, Michot B, Bachellerie J P. Nucleic Acids Res. 1983;11:5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker C T, Kloser A W, Schnaitman C A, Stein M A, Gottesman S, Gibson B W. J Bacteriol. 1992;174:2525–2538. doi: 10.1128/jb.174.8.2525-2538.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaara M. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caroff M, Karibian D. Appl Environ Microbiol. 1990;56:1957–1959. doi: 10.1128/aem.56.6.1957-1959.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caroff M, Deprun C, Karibian D. J Biol Chem. 1993;268:12321–12324. [PubMed] [Google Scholar]
- 21.Hall M N, Silhavy T J. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 22.Bonnefoy E, Almeida A, Rouviere-Yaniv J. Proc Natl Acad Sci USA. 1989;86:7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graeme-Cook K A, May G, Bremer E, Higgins C F. Mol Microbiol. 1989;3:1287–1294. doi: 10.1111/j.1365-2958.1989.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 25.Andersen J, Forst S A, Zhao K, Inouye M, Delihas N. J Biol Chem. 1989;264:17961–17970. [PubMed] [Google Scholar]
- 26.Mizuno T, Mizushima S. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 27.Mizuno T, Chou M Y, Inouye M. Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Painbeni E, Mouray E, Gottesman S, Rouviere-Yaniv J. J Mol Biol. 1992;234:1021–1037. doi: 10.1006/jmbi.1993.1656. [DOI] [PubMed] [Google Scholar]
- 29.Van Alphen W, Lugtenberg B. J Bacteriol. 1977;131:623–630. doi: 10.1128/jb.131.2.623-630.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balke V, Gralla J. J Bacteriol. 1987;169:4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tupper A E, Owen-Hughes T A, Ussery D W, Santos D S, Fergusson D J P, Sidebotham J M, Hinton J C D, Higgins C F. EMBO J. 1994;13:258–268. doi: 10.1002/j.1460-2075.1994.tb06256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nash H A, Robertson C. J Biol Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- 33.Huang L, Tsui P, Freudlich M. J Bacteriol. 1990;172:5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsui P, Helu V, Freudlich M. J Bacteriol. 1988;170:4950–4953. doi: 10.1128/jb.170.10.4950-4953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang K, Schieberl J L, Igo M M. J Bacteriol. 1994;176:1309–1315. doi: 10.1128/jb.176.5.1309-1315.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George A M, Levy B. J Bacteriol. 1983;155:531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seoane A S, Levy S B. J Bacteriol. 1995;177:530–535. doi: 10.1128/jb.177.3.530-535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]