Abstract
Almost all proteins mediating transcriptional activation from promoter-distal sites attach themselves, directly or indirectly, to specific DNA sequence elements. Nevertheless, a single instance of activation by a prokaryotic topologically linked DNA-tracking protein has also been demonstrated. The scope of the latter class of transcriptional activators is broadened in this work. Heterologous fusion proteins linking the transcriptional activation domain of herpes simplex virus VP16 protein to the sliding clamp protein β of the Escherichia coli DNA polymerase III holoenzyme are shown to function as topologically DNA-linked activators of yeast and Drosophila RNA polymerase II. The β:VP16 fusion proteins must be loaded onto DNA by the clamp-loading E. coli γ complex to be transcriptionally active, but they do not occupy fixed sites on the DNA. The DNA-loading sites of these activators have all the properties of enhancers: they can be inverted and their locations relative to the transcriptional start site are freely adjustable.
Regulation of transcription frequently involves cis-acting genomic sites that are located at some distance from the transcribed DNA and serve as assembly points for multiprotein regulatory complexes. It has been convincingly shown that some of these regulatory protein complexes convey their effects on transcription by making direct protein–protein contacts with the core transcription machinery, and it is generally assumed that transcriptional regulation by site-specific DNA-binding proteins is generated by such direct through-space interactions, with the connecting DNA looped out (1–6).
Other mechanisms of action can also be imagined (7), and prior work on the connection between DNA replication and expression of the late genes of bacteriophage T4 provides clear-cut evidence for a mechanism of transcriptional activation that is different in kind. Transcription of the T4 late genes is initiated at extremely simple promoters consisting of an 8-bp TATA box, but requires, in addition to the σ-family promoter recognition protein encoded by T4 gene 55 (gp55), a coactivator protein encoded by T4 gene 33 (gp33), and a transcriptional activator encoded by T4 gene 45 (gp45), acting through enhancer-like DNA-entry sites (8–10).
gp45 is the “sliding clamp” of the T4 replication machinery, serving to increase the processivity and speed of replicative DNA chain elongation (11–13). The homologs of gp45 in cellular DNA replication are the β subunit of Escherichia coli DNA polymerase III holoenzyme and, in eukaryotes, the proliferating cell nuclear antigen (PCNA) (14, 15). Each sliding clamp has the form of a ring with a central channel large enough to accommodate a DNA duplex; PCNA and gp45 are trimeric and β is a dimer (16, 17). Each protein tethers its conjugate DNA polymerase to the DNA template, and each sliding clamp requires a conjugate assembly factor (the “clamp loader”) acting at a suitable loading site, such as a nick or gap in double-stranded DNA or a double strand–single strand DNA junction. The specific loading factors of gp45 and β are the gp44/62 and γ complexes of the respective DNA polymerase holoenzymes. Loading of the sliding clamps onto DNA requires ATP hydrolysis; once they have been loaded, clamps slide freely along DNA by one-dimensional diffusion (14, 15, 18).
For gp45, the DNA-entry site functions as a transcriptional enhancer. Transcriptional activation requires a continuous and unobstructed path from the enhancer to the T4 late promoter, with which gp45 eventually becomes stably associated, situated at the upstream end of the transcription initiation complex, in close vicinity to, and interacting with, gp55 and gp33 (8, 9, 19, 20).
The preceding summary explains that proteins mediating transcriptional regulation from promoter-distal cis-acting sites are of two kinds: those that are physically fixed to specific DNA sequence elements and those that are topologically linked to DNA, but mobile in the one-dimensional space of the DNA thread. A “standard model” supported by diverse evidence describes the mode of action of a very large number of physically DNA-linked transcription-regulatory proteins (21–23). In contrast, only a single case of transcriptional activation by a topologically constrained DNA-tracking protein has thus far come to notice (20). We have addressed this imbalance by exploring the possibility of transcriptional activation of eukaryotic RNA polymerase by a topologically linked transcription factor. We show that chimeric proteins joining the transcriptional activation domain of herpes simplex virus VP16 to the E. coli sliding clamp β activate transcriptional initiation by the yeast (Saccharomyces cerevisiae) RNA polymerase II holoenzyme and by Drosophila melanogaster RNA polymerase II.
MATERIALS AND METHODS
Protein Production and Purification.
β–VP16 and VP16–β were overproduced in E. coli BL21 (DE3) harboring plasmid pLJ-1 and pLJ-12, respectively. pLJ-1 expresses a gene encoding the β protein fused to amino acids 413–490 of VP16 (24), and extended at the C end with the GSAWRHPQFGG streptavidin-affinity tag. pLJ-12 expresses a fusion gene encoding a protein with N-terminal sequence MHHHHHHPM followed by amino acids 413–490 of VP16, linked by a histidine residue to amino acids 1–366 of β. Complete sequences of these plasmids are available upon request. The streptavidin-tagged β–VP16 fusion protein was purified on a streptavidin-agarose affinity matrix and eluted with 5 mM diaminobiotin. His-tagged VP16–β was purified on a metal affinity column and eluted with 100 mM imidazole. These proteins were at least 95% pure, as judged by SDS/PAGE analysis.
Photochemical Crosslinking.
DNA tracking was assayed by photochemical crosslinking of partly double-stranded circular DNA (18, 25). For loading β–VP16 and β, the reaction mixture contained, in 15 μl, the specified quantities of β–VP16, β, γ complex (a generous gift from M. O’Donnell, Rockefeller University), 40 pmol of E. coli single-stranded DNA-binding protein (SSBE), and the specified concentrations of potassium acetate and polyethylene glycol (PEG; 3.3 kDa) in 33 mM Tris acetate, pH 7.8/10 mM magnesium acetate/0.8% (vol/vol) glycerol/250 μM dATP. Loading of VP16–β and β was done in the same reaction mixture, but with 100 mM Tris acetate, and without PEG. After incubation at 37°C for 15 min, samples were UV-irradiated, digested with nucleases, resolved by SDS/PAGE, and detected by phosphorimaging and autoradiography.
In Vitro Transcription Assays.
For transcription with the S. cerevisiae RNA polymerase II holoenzyme, each sample contained, in 25 μl, 200 ng of gpII-nicked DNA preloaded with β–VP16, as specified, 24 ng of TATA box-binding protein (TBP), 40 ng of transcription factor TFIIB (26), 3 ng of TFIIE (27), 3 μl of a crude TFIIH phosphocellulose fraction [prepared by M. S. Healy according to ref. 28] at 0.5 mg/ml total protein and 3 μl of a crude RNA polymerase II holoenzyme fraction (hydroxyapatite fraction of ref. 29) at 1 mg/ml total protein. DNA (≈0.09 pmol) was first assembled in 5 μl of 33 mM Tris acetate, pH 7.8/10 mM magnesium acetate/100 mM potassium acetate/0.8% (vol/vol) glycerol/10% (wt/vol) PEG (3.3 kDa) with the indicated amounts of β–VP16, γ complex, and 500 μM dATP for 15 min at 37°C, then added to 8 μl of a mixture containing RNA polymerase II holoenzyme, TBP, TFIIB, TFIIE, and TFIIH, and RNA synthesis was started by adding 12 μl of transcription buffer A (83 mM potassium Hepes, pH 7.8/16 mM magnesium acetate/3.2 mM dithiothreitol/1.67 mM ATP/1.67 mM CTP/0.1 mM UTP/10 μCi of [α-32P]UTP/0.2 unit of Prime RNase inhibitor; 1 μCi = 37 kBq). After 30 min at 23°C, transcription was terminated by adding 200 μl of stop mixture (10 mM Tris⋅HCl, pH 7.5/0.3 M NaCl/5 mM EDTA/0.1 mg/ml glycogen/12.5 units/ml RNase T1), and samples were processed as described (30).
For transcription with the D. melanogaster soluble nuclear extract (31), 5 μl of loading mixture containing 100 ng of template DNA, γ complex, VP16–β, and 1 mM ATP, preincubated at 37°C for 2–15 min, was mixed with 12.5 μl of transcription buffer B [12.5 mM potassium Hepes, pH 7.8/50 mM KCl/6.25 mM MgCl2/0.05 mM EDTA/5% (vol/vol) glycerol/0.5 mM dithiothreitol] and with 5 μl of soluble nuclear extract and histone H1 (32). After incubation at 25°C for 2–4 min, 3 μl of a solution containing 5 mM each of ATP, GTP, UTP, and CTP was added, and RNA synthesis at 25°C was terminated after 30 min. E4 transcripts were analyzed by primer extension.
RESULTS
Chimeric DNA-Tracking proteins.
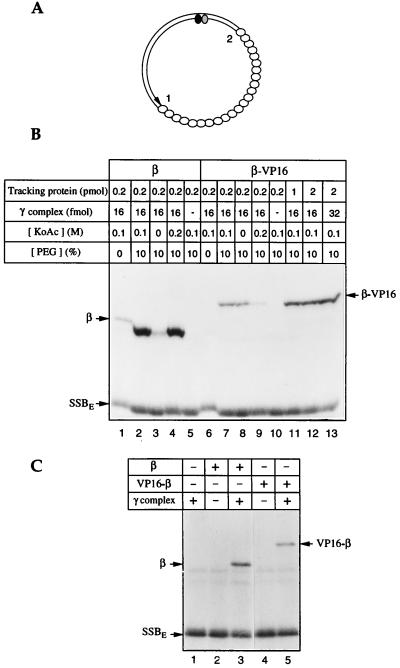
A topologically linked transcriptional activator modeled on the sliding clamps of the DNA polymerases must retain the ability to be loaded onto, and slide along, DNA (Fig. 1). The β:VP16 fusion proteins meet this criterion: attachment of the transcriptional activator domain of VP16 and affinity tags to the N or C terminus of β does not interfere with dimerization, as judged by size exclusion chromatography (data not shown). β interacts with its loading factor, the γ complex, through its C terminus (33); attaching the hoped-for transcriptional activator to this end of β does affect the ability to be loaded onto DNA. Nevertheless, as judged by photochemical cross-linking (18, 25) (Fig. 1B), β–VP16 can be loaded onto DNA in 10% PEG (3.3 kDa) (lanes 7, 8, and 11–13), although much less effectively than β (compare lanes 1–4 with lanes 6–9). Loading requires the γ complex (lanes 5 and 10) and dATP or ATP (data not shown). VP16–β (the fusion protein with the transcriptional activator domain joined to the N terminus of the sliding clamp) does not require macromolecular crowding for DNA loading, but it also is less efficiently loaded than is β (Fig. 1C).
Figure 1.
β–VP16 and VP16–β can be loaded onto DNA. β–VP16 and VP16–β fuse a transcriptional activation domain of VP16 (24) and affinity tags to the C and N ends of E. coli β, respectively. DNA loading and tracking were assessed by photochemical cross-linking (18, 25). (A) The photochemical probe contains a single residue of 5-[N′-(p-azidobenzoyl)-3-aminoallyl]-dUMP (ABdUMP) (shaded ellipse) next to the single radioactive nucleotide ([α-32P]dGMP) (filled ellipse) hundreds of base pairs from a preferred DNA-loading site 1 (a primer-template double strand–single strand junction) and ≈130 bp from a less efficient loading site 2 (the double strand–single strand junction of opposite polarity). The empty ellipses represent E. coli single-stranded DNA binding protein (SSBE) coating the single-stranded portion of the photoactive probe. (B) Loading β-VP16. Proteins, in quantities shown above each lane, were incubated with the photoactive DNA for 15 min at 37°C, in reaction buffer containing 250 μM dATP, and the specified concentrations of potassium acetate and polyethylene glycol. Tracking along DNA from the loading site to the vicinity of the photoactive nucleotide was detected by UV-irradiation, digestion of the resulting covalent protein-DNA adducts with nucleases, and resolution by SDS/PAGE, as described in ref. 25. (C) Loading of VP16–β. Tracking proteins (0.5 pmol of β or VP16–β) and/or γ complex (16 fmol) were incubated with the photoactive DNA for 15 min at 37°C, in reaction buffer containing 100 mM potassium acetate and 250 μM dATP. Samples were irradiated and processed as indicated above.
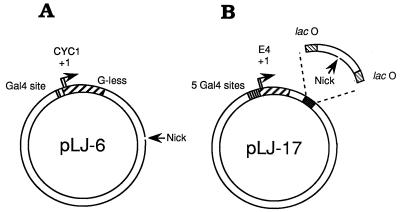
The activities of β–VP16 and of VP16–β as transcriptional effectors were examined in vitro. A form of yeast RNA polymerase II “holoenzyme” that requires supplementation with TFIIB, -E, and -H and TBP (29), but retains mediator activity [and is consequently responsive to transcriptional activators such as Gal4–VP16 (29, 34)], has been used to examine β–VP16. The transcription templates for these experiments (pLJ-5 and -6; Fig. 2A) incorporate a single Gal4-binding site, upstream of a transcription unit consisting of the CYC1 core promoter and a G-less cassette (29). pLJ-5 and -6 also have a site ≈900 bp downstream of the CYC1 transcriptional start sites for introducing a nick that serves as the DNA-loading site of β–VP16. In pLJ-5 the nick is in the transcribed strand of the CYC1 transcription unit, and in pLJ-6 it is in the nontranscribed strand (as shown in Fig. 2A), allowing β–VP16 to be loaded onto these two plasmids in opposite orientation (20). To examine activation of transcription by Drosophila RNA polymerase II, we constructed pLJ-17 to -20, each with five Gal4-binding sites upstream of the adenovirus E4 core promoter and a DNA-loading site for sliding clamps, inserted in either orientation and flanked by two symmetrical Lac operators (Fig. 2B).
Figure 2.
The transcription templates. (A) pLJ-6 contains the S. cerevisiae CYC1 promoter and a G-less cassette transcription unit. G-less transcripts initiating at CYC1 are ≈350 and ≈370 nt in length (35, 36). A DNA-loading site for β–VP16 is generated by cutting the nontemplate strand of this transcription unit with phage fd gpII endonuclease. In pLJ-5, the resulting DNA nick is located in the template strand. (B) pLJ-17 and -18 contain five Gal4-binding sites immediately upstream of the adenovirus E4 core promoter (base pairs −38 to +250 relative to the major transcriptional start site). The gpII nicking site is located between two symmetrical lac operators (37, 38), ≈390 bp from the RNA start site. The nick is in the transcribed strand for pLJ-17 and in the nontranscribed strand for pLJ-18. In plasmids pLJ-19 and -20, the nicking site is located ≈2 kbp from the RNA start site of the E4 promoter, in the transcribed strand for pLJ-19 and in the nontranscribed strand for pLJ-20. R3 Lac repressor, which forms a homodimer, and is, as a consequence, monovalent for DNA-binding (39, 40), was used to confine sliding clamps loaded at the single nick of pLJ-17 and -18 to an ≈100-bp DNA segment. Confinement was relieved by adding isopropyl β-d-thiogalactopyranoside (IPTG) (41, 42).
Transcriptional Activation of the Yeast Holoenzyme.
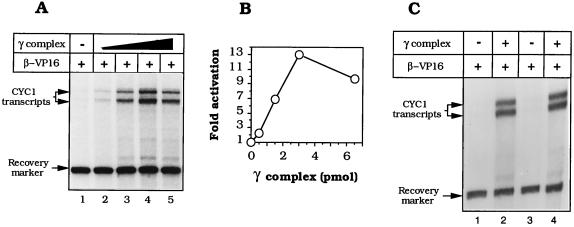
Assessing activation of transcription by RNA polymerase II in vitro requires an adequately low baseline of unactivated transcription: raising the electrolyte concentration (29) and diluting out the PEG that is required for adequate loading of β–VP16 (Fig. 1B) proved satisfactory for the yeast holoenzyme. Fig. 3 A and B shows an example of effective transcriptional activation by β–VP16 on nicked pLJ-6 DNA. These activation levels (7- to 13-fold) were approximately half that mediated by Gal4–VP16 when assayed under similar conditions (data not shown). Fig. 3C shows that transcription of pLJ-5 and -6 by the RNA polymerase II holoenzyme was comparably activated by β–VP16, indicating that there is no absolutely preferred orientation of β–VP16 on DNA relative to the transcription initiation complex at the CYC1 promoter. β–VP16 was ineffective without γ complex (Fig. 3A, lane 1), as was the γ complex in the absence of β–VP16 (not shown). Simply crowding the template with tracking β (43, 44) generated little or no activation and, as might be anticipated in view of the relatively inefficient loading of β–VP16, β interfered with transcriptional activation by β–VP16 (data not shown).
Figure 3.
β–VP16 activates transcription by yeast RNA polymerase II holoenzyme. The transcription reaction requires prior loading of the DNA-tracking protein onto the nicked template and subsequent dilution into the transcription medium. (A) Transcriptional activation by β–VP16. Reaction mixtures were assembled with 200 ng of nicked pLJ-6 DNA, 11 pmol of β–VP16, and 0, 0.5, 1.5, 3, or 6.5 pmol of γ complex (lanes 1–5) and assayed as outlined in the text. (B) Quantification of transcriptional activation, relative to basal transcription. (C) Transcription activation and polarity of loading. Reaction mixtures containing nicked pLJ-5 (lanes 1 and 2) or nicked pLJ-6 (lanes 3 and 4) were assembled with 11 pmol of β–VP16, without γ complex (lanes 1 and 3), or with 3.7 pmol of γ complex.
Activation of Drosophila RNA Polymerase II.
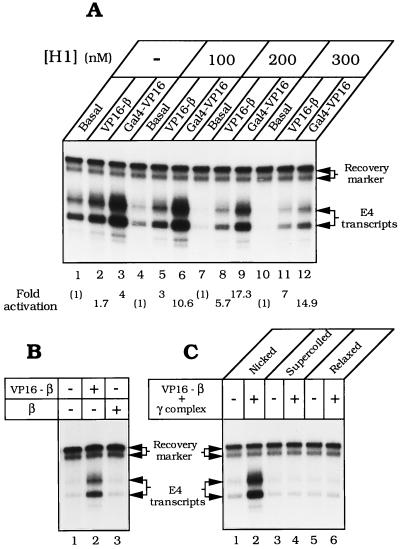
RNA polymerase II in the Drosophila embryo soluble nuclear extract responds to transcriptional activators bound to promoter-proximal sites (45). The ability to detect activation is increased greatly by adding histone H1, which depresses basal transcription (32). As shown in Fig. 4A, transcription of nicked pLJ-17 was weakly stimulated (≈1.5- to 4-fold) by Gal4–VP16 (lane 3) and responded even more weakly to VP16–β (lane 2). Addition of histone H1 progressively depressed basal transcription, exposing strong activation by VP16–β and by Gal4–VP16 (Fig. 4 A and B). Transcriptional activation required γ complex for loading VP16–β onto DNA (not shown), and β did not substitute for VP16–β in transcriptional activation (Fig. 4B, lane 3). Only nicked pLJ-17 served as a vehicle for transcriptional activation by the sliding clamp; supercoiled DNA and relaxed covalently closed DNA were unresponsive to VP16–β (Fig. 4C), although activated by Gal4–VP16 (not shown).
Figure 4.
Activation of transcription by Drosophila RNA polymerase II. (A) Efficient activation by VP16–β in the presence of histone H1. Reaction mixtures containing 100 ng of nicked pLJ-17 DNA were assembled with 6.25 pmol of VP16–β and 3.1 pmol of γ complex (lanes 2, 5, 8, and 11), or with 200 ng of Gal4–VP16 (lanes 3, 6, 9, and 12). (B) Activation (in presence of 200 nM histone H1) requires the VP16 activation domain. Reaction mixtures contained nicked pLJ-17 DNA, 3.1 pmol of γ complex, and 6.25 pmol of either VP16–β (lane 2) or β (lane 3). (C) Activation requires nicked DNA. Components specified at the top of each lane were assembled as described in the text. Transcripts were detected by primer extension and quantified by phosphorimaging. Gal4–VP16 was generously provided by E. Blackwood (University of California at San Diego) and R. Hori (University of California at Los Angeles).
The DNA-Loading Site of VP16–β Is an Enhancer.
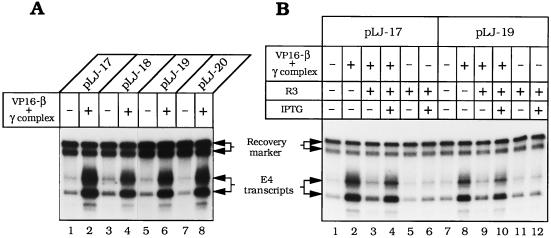
Constraints on the placement of the DNA-loading site for VP16–β were examined by constructing plasmids pLJ-18, -19, and -20. In pLJ-18, the DNA nick is in the nontranscribed strand of the E4 transcription unit, at approximately the same distance from the promoter (≈0.4 kbp) as in pLJ-17. In pLJ-19 and -20, the nick and the E4 promoter are almost at opposite poles of the 4.9-kbp plasmid, separated by ≈2 kbp of DNA. VP16–β activated transcription in all these plasmids to a comparable extent (Fig. 5A). The ability to reverse polarity and to be moved over long distances defines the DNA nick in these plasmids as the conjugate enhancer of VP16–β.
Figure 5.
VP16–β is a DNA-tracking transcriptional activator and its DNA-loading site is an enhancer. (A) Transcription of nicked pLJ-17 and -18 and pLJ-19 and -20 (Fig. 2) by Drosophila RNA polymerase II was examined in the presence or absence of 6.25 pmol of VP16–β and 3.1 pmol of γ complex, as specified above each lane (the autoradiographic exposure for lanes 1–4 is approximately half that for lanes 5–8). (B) Transcription of nicked pLJ-17 and -19 was examined in the presence of R3 Lac repressor (24.4 nM) in the absence (lanes 3, 5, 9, and 11) or in the presence of IPTG (20 μM) (lanes 4, 6, 10, and 12). R3 Lac repressor (40) was a generous gift from K. S. Mathews (Rice University). Other details are specified in the legend to Fig. 4.
When the bacteriophage T4 sliding clamp activates T4 late transcription, it requires an unobstructed DNA track between its loading site and the late promoter (20). The requirement for a clear track reflects the fact that the T4 sliding clamp exerts its effect on transcription as part of a compact promoter-bound initiation complex, in which it (gp45) touches at least two subunits of the T4-modified RNA polymerase holoenzyme (46). We examined whether transcriptional activation by VP16–β also requires an open DNA track between the enhancer and the promoter. When the R3 Lac repressor binds to the Lac operators flanking the DNA nick in pLJ-17 or -19, it confines VP16–β to an ≈100-bp segment of intervening DNA (Fig. 2B). R3 Lac repressor strongly diminished transcriptional activation by VP16–β but had no effect on basal transcription (Fig. 5B). IPTG, which detaches Lac repressor from its operator, substantially relieved the inhibition of activated transcription (in Fig. 5B, compare lanes 4, 3, and 2 with lanes 10, 9, and 8), but it had no effect on basal transcription (lanes 6 and 12). While nonspecific loading sites for loading VP16–β may eventually be generated by random nicking of DNA in this crude system, the effects of R3 Lac repressor and IPTG show that most of the transcriptional activation in this experiment derives from VP16–β loaded at the specific gpII nick.
In prior experiments on transcriptional activation in the presence of histone H1, diverse activators functioned from DNA-binding sites located within ≤300 bp of the promoter; activation at greater separations required nucleosomes as well as histone H1 (47, 48). Transcriptional activation in vitro in the presence of histone H1 only and with an enhancer that is separated from the promoter by 2 kbp (Fig. 5) is unprecedented, and it constitutes additional strong evidence that activation is primarily contributed by molecules of VP16–β that track along DNA from their intended loading site toward the promoter. The absence of polarity, manifested as equivalent transcriptional activation in pLJ-17 and -18 and in pLJ-19 and -20 (Fig. 5A) provides a striking contrast to transcriptional activation by the phage T4 gp45 sliding clamp, which has a strict requirement for a unique polarity of DNA loading (8, 9). This basic difference is probably due to the fact that the activation domain of VP16 is unstructured (24) and not rigidly connected to the β sliding clamp. Strict geometric constraints probably govern activation of the phage T4 late RNA polymerase holoenzyme (and perhaps bacterial RNA polymerases more generally); comparable constraints may be minimal for activated-when-touched (22, 23, 49, 50) eukaryotic RNA polymerase II.
DISCUSSION
These experiments suggest how topologically linked transcriptional activators might function in the context of eukaryotic transcription. The use of entirely heterologous components to construct the transcription systems emphasizes the potential generality of the activation mechanism. We constructed proteins with transcriptional activation domains covalently linked to the sliding clamp, but it should also be possible to fuse activators to noncovalent protein ligands of the sliding clamps, such as human p21 (51) or T4 phage gp55 (52). The ability to separate transcriptional activation from requirements for site-specific attachment to DNA should also open new possibilities for analyzing reaction pathways in initiation of transcription by RNA polymerase II.
It is appropriate to emphasize the distinction between DNA tracking by topologically linked proteins and DNA scanning by DNA-binding domains of proteins that are not topologically confined to it. DNA tracking by β–VP16 and VP16–β fusion proteins requires no affinity for DNA whatsoever; their range is limited only by the one-dimensional diffusion constant and the stability of the topological linkage. On the other hand, scanning of nonspecific DNA sequences by DNA-binding proteins, such as Lac repressor, depends on lateral transfer between weak binding sites and, unless sustained by additional DNA valences that support a kind of brachiation, is characteristically short range (53–56).
It will, of course, be appropriate and desirable to extend this line of experiments to include chromatin templates. However, the work that is presented here does serve as a demonstration of concept. Moreover, using nonnucleosomal DNA templates has made it possible to demonstrate unambiguously that transcriptional activation by the sliding clamps depends on DNA tracking. That will be (at best) difficult to prove in the context of chromatin because of DNA compaction and the consequent potentiation of activation through space. Be that as it may, the following considerations lead us to expect that topologically linked transcriptional activators can be made to function in the context of eukaryotic chromatin: (i) Nucleosomes are not fixed on DNA but mobile (57–59). Thus, although the rate of sliding of topologically linked proteins may be considerably limited by chromatin, the continuous action of chromatin remodeling factors should prevent sliding clamps from being completely immobilized by nucleosomes. (ii) Proteins that track along DNA should be capable of activating eukaryotic transcription through space, just as site-specifically bound activators are thought to do. Chromatin may extend the range and effect of topologically linked and physically bound transcriptional activators by compacting and organizing the intervening DNA. (iii) The RNA polymerase holoenzyme may carry its own nucleosome disruption machinery, although that issue is not entirely resolved (60, 61).
One can see no reason why the participation of topologically linked and physically bound transcriptional activators should necessarily be mutually exclusive; rather, it should be possible for them to act in concert. Harnessing transcriptional activation by topologically linked proteins to gene-specific action should also be possible. Regionally and temporally confined action could, for example, be secured by regulating the synthesis of the transcriptional activator, specifying the DNA-loading site with a sequence-specific endonuclease, and limiting the sliding range of the activator on DNA with boundary elements.
Acknowledgments
We are grateful to E. Blackwood, L. DeVito, M. S. Healy, R. Hori, K. S. Matthews, and M. O’Donnell for generous gifts of material; J.-P. Léonetti, A. Kumar, G. A. Kassavetis, and R. L. Tinker-Kulberg for their interest in this work and advice; and M. Filutowicz and P. H. von Hippel for helpful comments on the manuscript. Our research has been supported by grants (to M.H.S., J.T.K. and E.P.G.) from the National Institute of General Medical Sciences and to J.T.K. from the National Science Foundation. M.H.S. gratefully acknowledges a Junior Faculty Research Award from the American Cancer Society and J.T.K. is a Presidential Faculty Fellow.
ABBREVIATIONS
- TBP
TATA box-binding protein
- TF
transcription factor
- IPTG
isopropyl β-d-thiogalactopyranoside
References
- 1.Wedel A, Weiss D S, Popham D, Dröge P, Kustu S. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 2.Su W, Porter S, Kustu S, Echols H. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ptashne M, Gann A A. Nature (London) 1990;346:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 4.Green M R. Harvey Lect. 1993;88:67–96. [PubMed] [Google Scholar]
- 5.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 6.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 7.Wang J C, Giaever G N. Science. 1988;240:300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- 8.Herendeen D R, Kassavetis G A, Barry J, Alberts B M, Geiduschek E P. Science. 1989;245:952–958. doi: 10.1126/science.2672335. [DOI] [PubMed] [Google Scholar]
- 9.Herendeen D R, Williams K P, Kassavetis G A, Geiduschek E P. Science. 1990;248:573–378. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- 10.Brody E N, Kassavetis G A, Ouhammouch M, Sanders G M, Tinker R L, Geiduschek E P. FEMS Microbiol Lett. 1995;128:1–8. doi: 10.1111/j.1574-6968.1995.tb07491.x. [DOI] [PubMed] [Google Scholar]
- 11.Nossal N. In: Molecular Biology of Bacteriophage T4. Karam J D, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 43–55. [Google Scholar]
- 12.Kaboord B F, Benkovic S J. Biochemistry. 1996;35:1084–1092. doi: 10.1021/bi9520747. [DOI] [PubMed] [Google Scholar]
- 13.Young M C, Weitzel S E, von Hippel P H. J Mol Biol. 1996;264:440–452. doi: 10.1006/jmbi.1996.0652. [DOI] [PubMed] [Google Scholar]
- 14.Stukenberg P T, Studwell-Vaughan P S, O’Donnell M. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 15.Kelman Z, O’Donnell M. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 16.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 17.Krishna T S, Kong X P, Gary S, Burgers P M, Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 18.Tinker R L, Kassavetis G A, Geiduschek E P. EMBO J. 1994;13:5330–5337. doi: 10.1002/j.1460-2075.1994.tb06867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tinker R L, Williams K P, Kassavetis G A, Geiduschek E P. Cell. 1994;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 20.Herendeen D R, Kassavetis G A, Geiduschek E P. Science. 1992;256:1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- 21.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D, editors. Molecular Biology of the Cell. New York: Garland; 1994. [Google Scholar]
- 22.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 23.Koleske A J, Young R A. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 24.Triezenberg S J. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 25.Bartholomew B, Tinker R L, Kassavetis G A, Geiduschek E P. Methods Enzymol. 1995;262:476–494. doi: 10.1016/0076-6879(95)62039-7. [DOI] [PubMed] [Google Scholar]
- 26.Feaver W J, Henry N L, Bushnell D A, Sayre M H, Brickner J H, Gileadi O, Kornberg R D. J Biol Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 27.Sayre M H, Tschochner H, Kornberg R D. J Biol Chem. 1992;267:23383–23387. [PubMed] [Google Scholar]
- 28.Svejstrup J Q, Feaver W J, LaPointe J, Kornberg R D. J Biol Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- 29.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 30.Sayre M H, Tschochner H, Kornberg R D. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 31.Kamakaka R T, Kadonaga J T. Methods Cell Biol. 1994;44:225–235. doi: 10.1016/s0091-679x(08)60916-4. [DOI] [PubMed] [Google Scholar]
- 32.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 33.Naktinis V, Turner J, O’Donnell M. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 34.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 35.Sawadogo M, Roeder R G. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lue N F, Flanagan P M, Sugimoto K, Kornberg R D. Science. 1989;246:661–664. doi: 10.1126/science.2510298. [DOI] [PubMed] [Google Scholar]
- 37.Sadler J R, Sasmor H, Betz J L. Proc Natl Acad Sci USA. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simons A, Tils D, von Wilcken-Bergmann B, Müller-Hill B. Proc Natl Acad Sci USA. 1984;81:1624–1628. doi: 10.1073/pnas.81.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alberti S, Oehler S, von Wilcken-Bergmann B, Müller-Hill B. EMBO J. 1993;12:3227–3236. doi: 10.1002/j.1460-2075.1993.tb05992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Alberti S, Matthews K S. J Biol Chem. 1994;269:12482–12487. [PubMed] [Google Scholar]
- 41.Gilbert W, Müller-Hill B. Proc Natl Acad Sci USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riggs A D, Bourgeois S. Biophys J. 1969;9:A84. [Google Scholar]
- 43.Fu T-J, Sanders G M, O’Donnell M, Geiduschek E P. EMBO J. 1996;15:4414–4422. [PMC free article] [PubMed] [Google Scholar]
- 44.Yao N, Turner J, Kelman Z, Stukenberg P T, Pan Z-Q, Hurwitz J, O’Donnell M. Genes to Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 45.Kamakaka R T, Tyree C M, Kadonaga J T. Proc Natl Acad Sci USA. 1991;88:1024–1028. doi: 10.1073/pnas.88.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders, G. M., Kassavetis, G. A. & Geiduschek, E. P. (1997) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 47.Croston G E, Laybourn P J, Paranjape S M, Kadonaga J T. Genes Dev. 1992;6:2270–2281. doi: 10.1101/gad.6.12a.2270. [DOI] [PubMed] [Google Scholar]
- 48.Laybourn P J, Kadonaga J T. Science. 1992;257:1682–1685. doi: 10.1126/science.1388287. [DOI] [PubMed] [Google Scholar]
- 49.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 50.Stargell L A, Struhl K. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 51.Gulbis J M, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 52.Tinker-Kulberg R L, Fu T-J, Geiduschek E P, Kassavetis G A. EMBO J. 1996;15:5032–5039. [PMC free article] [PubMed] [Google Scholar]
- 53.von Hippel P H, Berg O C. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 54.Ruusala T, Crothers D M. Proc Natl Acad Sci USA. 1992;89:4903–4907. doi: 10.1073/pnas.89.11.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson S A, Glass R E, Shimamoto N. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 56.Surby M A, Reich N O. Biochemistry. 1996;35:2201–2208. doi: 10.1021/bi951883n. [DOI] [PubMed] [Google Scholar]
- 57.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 58.Imbalzano A N, Schnitzler G R, Kingston R E. J Biol Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 59.Pazin M J, Bhargava P, Geiduschek E P, Kadonaga J T. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- 60.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 61.Cairns B R, Lorch Y, Li Y, Zhang M C, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]