Abstract
Abalone sperm lysin is a 16-kDa acrosomal protein, which nonenzymatically and species selectively creates a hole in the egg vitelline envelope (VE) through which the sperm passes to reach the egg cell membrane. The crystal structures of both monomeric and dimeric lysins have been solved and the sequences of lysins from 20 abalone species have been determined. As a first step in understanding the molecular mechanism by which lysin creates a hole in the VE, its VE receptor was isolated. The VE receptor for lysin (VERL) is an unbranched, rod-like molecule with an approximate relative molecular mass of 2 million; half the mass being carbohydrate. Fluorescence polarization studies showed positive cooperativity in the binding of lysin to VERL (EC50 ≈9 nM) and were consistent with the species selectivity of lysin in dissolving VEs. Each molecule of VERL bound between 126 and 142 molecules of monomeric lysin (two independent assays), showing that VERL possesses repetitive lysin-binding motifs.
Many marine invertebrate species spawn gametes into seawater, where fertilization and embryogenesis occur. In sea urchins (1, 2) and abalone (3, 4), several closely related, congeneric species may have overlapping breeding seasons and habitats. Such species usually exhibit species selectivity (specificity) in experimental cross-fertilizations, meaning that fertilization occurs more readily between homospecific mixtures of sperm and eggs than between heterospecific mixtures. The demonstrated species selectivity in the recognition events between invertebrate sperm and egg is a valuable model bearing on the general problem of the biochemistry of molecular recognition between cells.
In marine invertebrates, the study of gamete recognition proteins may also yield information on the mechanism by which species arise in the sea, where barriers to larval transport are not readily apparent (5). Evidence that gamete recognition proteins may be involved in speciation comes from analyses of nucleotide substitutions in sea urchin sperm bindins (1) and abalone sperm acrosomal proteins (4, 6). Such analyses show that the evolution of these proteins has been promoted by positive Darwinian selection, suggesting there is significant adaptive value in altering their primary structure.
The availability of large quantities of marine invertebrate gametes permits a detailed characterization of their gamete surface molecules involved in fertilization. The abalone egg is surrounded by an elevated, glycoproteinaceous vitelline envelope (VE) 0.6 μm in diameter (7, 8). Spermatozoa bind to the VE, their acrosomal granule exocytoses, and lysin is released. Lysin creates a 3-μm diameter hole in the VE through which the spermatozoan passes to reach the egg cell membrane. Electron micrographs of the lysin-created hole show it is composed of splayed-apart fibers of 13-nm diameter; covalent bonds are not broken when lysin disrupts the VE (7).
Although gamete recognition proteins have been cloned from both mammals (9–13) and invertebrates (1–4, 6, 14–16), the molecular mechanism of action of all these proteins remains unknown. Abalone sperm lysin is the only gamete recognition protein whose crystal structure is known (17). There are three important features of the monomeric crystal structure: a species-unique amino-terminal domain of residues 1–12 projects away from the α-helical bundle comprising the remainder of the molecule (residues 13–136); one surface of the monomer has two tracks of basic residues along its entire length; and there is a surface-exposed patch of 11 hydrophobic residues on the opposite surface from the basic tracks (17). The basic tracks and hydrophobic patch residues of lysins of the seven species of California abalone are conserved, suggesting they are important in the mechanism of VE disruption, but not in the mechanism of species recognition (18). Lysin monomers dimerize (approximate KD = 1 μM) by the interaction of hydrophobic patches. Addition of isolated VEs causes the rapid dissociation of dimers and the binding of monomeric lysin to the VE (19).
Having obtained structural, biochemical, and evolutionary information about abalone sperm lysin, we proceeded to identify, isolate and characterize the receptor (ligand) in the VE to which lysin binds. Our goal is 2-fold: first, to describe the molecular mechanism of species-selective recognition between lysin and the VE, and second, to determine the molecular mechanism by which lysin nonenzymatically disrupts the VE.
MATERIALS AND METHODS
Affinity Purification of VE Receptor for Lysin (VERL).
Five milligrams of CM-cellulose purified (3) Haliotis rufescens (Hr) or Haliotis corrugata (Hc) sperm lysin was coupled to 1 ml Affi-Gel 10 (Bio-Rad) in 10 mM Mes (pH 6.0)/500 mM NaCl, following the manufacturer’s instructions. The Affi-Gel 10 was blocked with ethanolamine and washed with 10 mM Hepes buffered seawater, pH 7.8 (HSW). VEs were isolated from unfertilized abalone eggs as described (7) and dissolved by a 2-min exposure to pH 2.8 seawater (adjusted with acetic acid). The pH was readjusted to 7.8 with 1 M Tris (pH 8) and the mixture centrifuged for 2 min at 5,000 × g. The resulting supernatant is referred to throughout as “soluble VE material.” This material was applied to the lysin–Affi-Gel 10, the columns were washed with 50 ml HSW, eluted using 1 ml of 100 mM glycine (pH 2.8), and the eluate immediately neutralized with 200 μl of 1 M Tris, pH 8.0. Two control columns, one of ethanolamine-blocked Affi-Gel alone and the other coupled with 5 mg lysozyme (a similar pI and size as lysin) did not bind soluble VE material.
Sucrose Density Gradient Centrifugation.
Purified lysin was radioiodinated as described (19) and added to soluble VE material and the mixture layered on a 13 ml 5–55% sucrose gradient in HSW, which was centrifuged at 200,000 × g for 20 h in a Beckman SW41 rotor (4°C). Seventeen fractions were taken and portions analyzed by SDS/PAGE and autoradiography. Radioiodination does not alter the ability of lysin to disrupt VEs (19). Control gradients of lysin alone without soluble VE material were also run; following centrifugation, lysin remained in the top two fractions.
VE Dissolution Assays.
Isolated egg VEs (7) were suspended in filtered seawater containing 1 mg/ml bovine serum albumin, 10 mM sodium azide, 10 mM Tris (pH 8.0), and 600 μl placed in a 1 ml cuvette. One hundred microliters of seawater containing various amounts of purified lysin (3) was added and the decrease in turbidity with time measured at 640 nm (23°C; refs. 3 and 7). To test if VERL fractions inhibited the activity of lysin, varying amounts of VERL were added to 6.5 μg lysin and the 100 μl volume incubated for 30 min at 23°C before addition to the 600 μl of VE suspension in a 1 ml cuvette.
Polyacrylamide Gel Electrophoresis (PAGE).
The recipes of Laemmli (20) were used for slab gel electrophoresis in SDS and mercaptoethanol. Samples were boiled for 5 min in sample buffer (20) before loading. For 2.5% gels, the acrylamide:bisacrylamide weight ratio was 38:1. For 2.5–15% gradient gels, equal volumes of each concentration of acrylamide were used. Rabbit muscle acetone powder (Sigma) containing titin (2,800 kDa), nebulin (770 kDa), and myosin (205 kDa) was used for relative molecular mass standards (21). Gels were qualitatively (22) or quantitatively silver stained and used for densitometry (23).
Electron Microscopy.
Sucrose gradient fractions containing Hr VERL were dialyzed into HSW containing 10 mM sodium azide. Formvar-coated, carbon-backed grids were dipped into the dialysate, blotted, exposed to 1% ultra-filtered uranyl acetate, blotted on edge, air dried, and viewed by transmission electron microscopy.
Peptide Mapping.
Hc VERL was isolated by lysin–affinity chromatography, 125I-labeled (19), and electrophoresed on SDS/2.5% polyacrylamide gels. The three VERL bands were detected by autoradiography and electroeluted using a Centriluter device (Amicon). Eluted samples were boiled for 5 min and digested for 1 h at 37°C with 1.3 μg V8 Endoproteinase Glu-C (Sigma). Digests were separated on a reducing SDS/12.5% polyacrylamide gel and autoradiograms prepared.
Fluorescence Polarization.
Lysin was fluorescently labeled (19) with tetramethyl rhodamine isothiocyanate (Molecular Probes) to a dye:protein molar ratio of 0.3. A 1 μM solution of labeled lysin in HSW was titrated with increasing quantities of VERL (protein mass estimated as 1 million). Anisotropy (r) was measured 10 times at each point (excitation 547 nm, emission 572 nm, bandpass 5 nm, 5 s integration, G factor = 0.632) on a SPEX Fluoromax-2 spectrofluorimeter. Each point is an average of three experiments. Data were analyzed using nonlinear regression (SigmaPlot, Jandel, San Rafael, CA) with the equation Δr = Δrmax [VERL]n/((EC50)n + [VERL]n), where Δr = robserved − rinitial, Δrmax = rmax − rinitial, n = Hill coefficient, EC50 = effective concentration of half saturation, and [VERL] is the concentration of VERL added. Values of Δrmax, EC50, and n were used as fitting parameters. The values obtained by curve fitting are similar to those obtained by independent means (Δrmax was experimentally determined, while EC50 and n were determined from Hill plots). All plots fit the equation with r2 > 0.99. Data were plotted as the fraction bound (Δr/Δrmax) against nanomoles of total VERL. Although the concentration of VERL, compared with lysin, is low in these experiments, the high stoichiometry observed indicates that the concentration of VERL–lysin binding sites is high. Experiments with 0.1 μM lysin yielded similar results to 1.0 μM lysin.
Lysin–VERL Stoichiometry.
Sedimentation assay. Lysin was 125I-labeled to 230,000 cpm/μg. Fifteen microgram-labeled lysin was mixed with varying amounts of VERL [1.5 μg (1.5 pMol) to 3 μg (3 pMol)] in a total volume of 100 μl HSW containing 2 mg/ml BSA and 10 mM sodium azide in a 400-μl Beckman Airfuge tube, which was overlaid with 300 μl of mineral oil. After 30 min, the tubes were centrifuged in an Airfuge at 180,000 × g for 12 h (23°C). Ten microliter portions of the aqueous phase supernatant were counted in triplicate and the depletion of labeled lysin determined. Control tubes of VERL without lysin showed all the VERL pelleted.
Fluorescence polarization assay.
The fluorescence anisotropy of a 10.5 μM solution of rhodamine-labeled lysin (19) was measured after addition of varying amounts of VERL [1.5 μg (1.5 pMol) to 15 μg (15 pMol)] and after 1 min the anisotropy was measured. The change in anisotropy was plotted against the lysin:VERL molar ratio and the inflection point taken as the maximum binding stoichiometry (24).
Miscellaneous Procedures.
Gel filtration chromatography of soluble VE material was done in HSW containing 10 mM sodium azide on a 1 × 30 cm column of Sephacryl S-500 (working range for dextrans, 40,000 to 20 million molecular mass). Protein was determined by the bicinchoninic acid assay (Pierce) with BSA as standard. Hexose (total carbohydrate) was assayed by the phenol-sulfuric acid method (25) with fucose as standard.
Qualitative carbohydrate composition was done by the University of California, San Diego Glycobiology Core Facility on samples of VERL lyophilized from distilled water. Amino acid compositional analyses of soluble VE material (representing all the molecules of VEs) and VERL isolated on sucrose density gradients, were done by the Stanford University Protein and Nucleic Acid Facility.
RESULTS
Identification and Isolation of VERL.
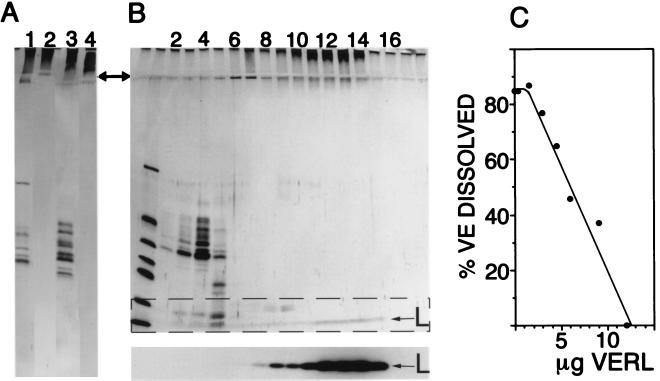
VERL was identified by two methods: affinity chromatography using lysin–Affi-Gel columns and sucrose density gradient sedimentation. Fig. 1A shows the affinity purification of VERL from two species of abalone Hc (lanes 1 and 2) and Hr (lanes 3 and 4). Lanes 1 and 3 show that the pH 2.8-soluble VE material applied to the lysin–Affi-Gel is composed of fewer than 10 silver-staining components, most ranging in relative mass from 30–50 kDa, and high molecular mass material remaining in the stacking gel. After extensive washing of the columns with HSW, the only material eluting in pH 2.8 glycine is this high molecular mass material remaining in the stacking gel (lanes 2 and 4).
Figure 1.
Identification of VERL. (A) Affinity purification of VERL from two abalone species (Hc and Hr). Lanes: 1, total pH 2.8-soluble VE material (Hc); 2, material eluted by pH 2.8 glycine from the Hc lysin column; 3, starting material from Hr VEs; 4, pH 2.8-eluted material from the Hr lysin column. Material eluted from lysin columns by low pH does not enter the 12% separating gel (arrow notes interface between stacking and separating gels). (B) Sucrose density gradient sedimentation of lysin bound to VERL. (Upper) Silver-stained SDS/12% polyacrylamide gel with the gradient fractions from top (light) to bottom (heavy) going left to right. Lanes: 1, molecular mass standards (13–67 kDa); 2–5, VE components ranging from 30–50 kDa; 9–14, high molecular mass VE material not entering the separating gel. (Lower) Autoradiogram of the dashed rectangle in Upper: 125I-lysin (L) comigrates with the material that does not enter the separating gel. In the absence of VE material, 125I-lysin remained in the top two fractions (not shown). (C) Isolated VERL inhibits the ability of lysin to dissolve isolated VEs. Fifty percent inhibition occurred at a lysin:VERL molar ratio of 69:1.
Total soluble VE material (Hc species) was mixed with 3.2 × 106 cpm radioiodinated Hc lysin (77,000 cpm/μg) and the mixture subjected to sucrose density gradient centrifugation. Thirty-microliter portions of the gradient fractions (700 μl fractions) were analyzed by SDS/12% PAGE and autoradiography of the dried, silver-stained gels. Fig. 1B shows that the 125I-lysin comigrates with the material remaining in the stacking gel (fractions 9–14). This high molecular mass material was recovered and found to inhibit the ability of lysin to dissolve VEs (Fig. 1C). Fifty percent inhibition occurred at a lysin: VERL molar ratio of 69:1 (based on a protein mass estimate for VERL of 1 million).
Electrophoretic Analysis of VERL.
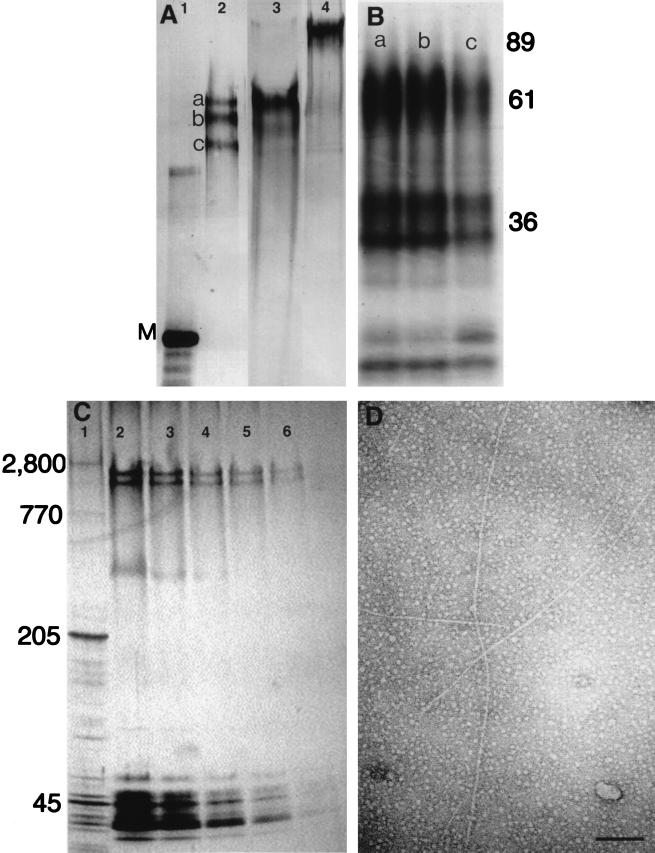
VERL, isolated by lysin–affinity chromatography, was analyzed on SDS/2.5% polyacrylamide gels (Fig. 2A) on which it separated into 1–3 components with relative molecular masses ranging from 1,500–2,500 kDa. In the absence of mercaptoethanol, Hr VERL appeared to be of larger size, suggesting that it might be a disulfide bonded dimer, or that disulfide bonds may be involved in its native structure. One-dimensional peptide maps of the three bands shown in Fig. 2A (lane 2) showed that the three VERL bands had identical digestion patterns, indicating that all three have the same protein backbone (Fig. 2B, lanes 1–3).
Figure 2.
Characterization of VERL purified by lysin–affinity chromatography. (A) Silver-stained SDS/2.5% polyacrylamide gel. Lanes: 1, rabbit myosin (M; 205 kDa); 2, Hc VERL resolved as three components (a–c) under reducing conditions; 3, Hr VERL resolved as one component under reducing conditions; 4, Hr VERL under nonreducing conditions. (B) Autoradiogram of a SDS/12% polyacrylamide gel of Endoproteinase Glu-C-generated one-dimensional peptide maps of the three Hc VERL bands shown in A (a–c). Positions of molecular mass standards shown on right. (C) A SDS/2.5–15% polyacrylamide gradient gel of isolated Hr VEs quantitatively silver stained (23) and used for densitometry. Lanes: 1, rabbit muscle acetone powder with titin (2,800 kDa) and nebulin (770 kDa) as relative mass standards; 2–6, 50% dilutions of Hr VEs. VERL resolves as two bands below titin. Densitometry of these lanes showed that VERL was approximately 30% of the protein silver-staining material. (D) Isolated Hr VERL negatively stained with uranyl acetate and viewed by transmission electron microscopy. The VERL rods are approximately 13 nm in diameter (7). (Scale bar = 100 nm.)
To determine the relative abundance of VERL in the VE, isolated VEs were separated on gradient gels at different protein loads (Fig. 2C), which were silver stained and analyzed by densitometry. Using these techniques, VERL represented approximately 30% of the protein stain.
Electron Microscopy and Compositional Analysis.
Samples of Hr VERL, isolated by sucrose density gradient sedimentation, were viewed in the electron microscope after negative staining with uranyl acetate. The micrographs show that VERL is a long, unbranched rod of 13 nm diameter (7, 8). The size distribution of lengths was not determined, however, the 20 longest rods imaged were 630 ± 170 nm. The large size of VERL was confirmed by the finding that it eluted from a Sephacryl S-500 column close to the void volume, indicating a probable molecular mass of >1 million (data not shown).
Compositional assays of Hc and Hr VERL yielded estimates of 50% protein:50% carbohydrate. Carbohydrate analysis indicated that Hc VERL, isolated by lysin–affinity chromatography, is 16% fucose, 52% glucose, 20% mannose, 7% galactosamine, and 5% glucosamine. Amino acid compositional analysis of Hc and Hr VERLs shows them to differ considerably from total soluble VE material representing all components of the VE. VERLs are rich in Asp, Glu, Pro, and Ser, whereas soluble VE material is rich in Thr (Table 1).
Table 1.
Amino acid composition of VERL and VEs from abalone species Hc and Hr
| Amino acid | VERL, mol %
|
VEs, mol %
|
||
|---|---|---|---|---|
| Hc | Hr | Hc | Hr | |
| Glu | 10.6 | 10.2 | 8.0 | 8.0 |
| Thr | 9.5 | 9.3 | 28.0 | 29.2 |
| Ser | 13.1 | 10.8 | 8.2 | 7.7 |
| Asp | 11.4 | 11.4 | 6.4 | 5.9 |
| Pro | 11.4 | 12.2 | 8.6 | 8.5 |
| Gly | 5.0 | 4.9 | 6.6 | 6.9 |
| Ala | 5.6 | 5.3 | 5.1 | 5.2 |
| Val | 6.8 | 6.7 | 5.2 | 4.8 |
| Met | 0.3 | 0.6 | 0.7 | 0.5 |
| Ile | 4.5 | 4.8 | 3.1 | 3.2 |
| Leu | 4.8 | 6.7 | 4.8 | 4.9 |
| Tyr | 5.8 | 5.3 | 3.1 | 2.8 |
| Phe | 1.5 | 2.1 | 2.3 | 2.4 |
| His | 0.9 | 1.2 | 1.1 | 1.2 |
| Lys | 6.5 | 6.0 | 5.9 | 5.8 |
| Arg | 2.1 | 2.3 | 3.0 | 3.0 |
Cys and Trp not determined.
Fluorescence Polarization.
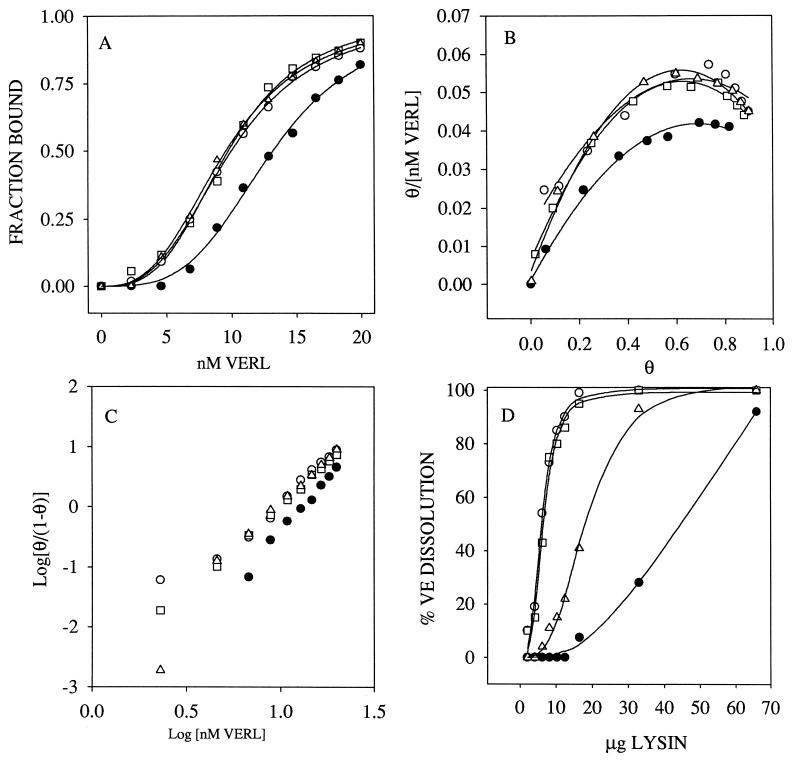
The interaction of lysin and VERL from Hc and Hr species was studied by fluorescence polarization. Sigmoidal binding curves were obtained indicating positive cooperativity in the interaction of these two gamete components (Fig. 3A). Positive cooperativity is supported by convex Scatchard plots (Fig. 3B), Hill plots with a slope of approximately three (Fig. 3C), and the fractional occupancy increasing from 10–90% over 1.1 log units of VERL concentration (Fig. 3A). Three of the binding curves are nearly identical, with effective concentrations of half saturation (EC50) ranging from 9.54–9.96 nM and Hill coefficients of 2.88–3.17. In the mixture of Hr lysin and Hc VERL, the curve is displaced, with an EC50 of 13.17 nM and a Hill coefficient of 3.50. In this heterospecific mixture, more Hr lysin must be added before binding to VERL commences. However, once binding of lysin to VERL begins, the ascending slopes are approximately parallel in all combinations of these two species. The species selectivity observed in these binding curves agrees with the data on lysin-mediated disruption of isolated VEs (Fig. 3D).
Figure 3.
Lysin–VERL binding is consistent with lysin-mediated VE dissolution. (A) Fluorescence polarization of VERL–lysin binding. The sigmoidal binding curves indicate positive cooperativity, as supported by convex Scatchard plots (B) and Hill plots with slopes of approximately 3 (C). (B and C) θ = the fraction of lysin bound. (D) Lysin-mediated VE dissolution measured by light scattering. Species combinations are: □, Hr VERL + Hr lysin; ○, Hc VERL + Hc lysin; ▵, Hr VERL + Hc lysin; •, Hc VERL + Hr lysin. The species selectivity in lysin–VERL binding curves (A) is consistent with the species selectivity of lysin in dissolving isolated VEs (D).
Estimates of the Number of Lysins Bound per VERL.
The large apparent size of VERL compared with the relatively small size of lysin (16 kDa), and the finding that the binding of lysin to VERL exhibits positive cooperativity, suggest that multiple lysins bind each VERL. A stoichiometric binding assay using fluorescent lysin and fluorescence polarization yielded a molar estimate of 142 ± 18 (n = 7) lysin monomers bound per VERL. Alternatively, an ultracentrifuge (Airfuge) sedimentation assay using 125I-lysin, yielded a stoichiometry of 126 ± 87 (n = 8) lysin monomers bound per VERL.
DISCUSSION
Egg Envelope Molecules Interacting with Sperm.
Most unfertilized animal eggs are surrounded by a glycoproteinaceous coat or envelope. There are two basic types of such structures containing proteins that interact with sperm during fertilization. The first type is intimately bonded to the egg plasma membrane and is most extensively characterized in the sea urchin (14, 15, 26). The second type of egg envelope is elevated from the egg plasma membrane and is found in mollusks (7, 8, 27), ascidians (28–30), amphibians (31, 32), and mammals (9, 33, 34). This discussion will pertain only to envelopes that are elevated from the plasma membrane and that the sperm must penetrate to reach the egg plasma membrane.
The mouse and the abalone are the only animals with elevated egg envelopes where the interacting components from both gametes have been isolated and their interaction studied in vitro. The envelope of the mouse egg, the zona pellucida (ZP), is composed of three glycoproteins. One of these molecules, ZP3, is the species-specific ligand (33) for the sperm membrane protein sp56 (12). Binding of ZP3 to sp56 induces the sperm acrosome reaction, which allows the sperm to pass through the ZP by a mechanism that probably involves proteolysis. The binding of ZP3 to sperm is complex and displays positive cooperativity (35).
Solubility studies on abalone egg VEs support the hypothesis that its structural integrity derives from hydrogen bonding among glycoprotein fibers (unpublished data). The disruption of the VE may occur by lysin stereospecifically severing inter-VE hydrogen bonds. The structure of lysin is consistent with such a hypothesis, which would explain its nonenzymatic action and tight binding to VE molecules (19). Knowledge of the structure of VERL may resolve how it and lysin interact species specifically in the disruption of the VE.
In studying the proteins of animal egg envelopes that interact with sperm, one potential problem is that large molecules such as VERL might be overlooked because they do not enter regularly formulated SDS/polyacrylamide gels. For example, the original report of the macromolecular components of abalone VEs showed considerable Coomassie blue staining material at the top of the stacker gel, which was overlooked as being of importance (ref. 7, figure 10).
Evidence That VERL Is the Receptor for Lysin.
The high molecular mass material in isolated VEs was identified as the receptor for lysin (VERL) on the basis of the following criteria: (i) it was the only component of VEs binding to lysin–Affi-Gel columns (Fig. 1A), (ii) lysin cosedimented with this material by sucrose density gradient sedimentation (Fig. 1B), (iii) fractions of this high molecular mass material inhibited lysin’s dissolution of isolated VEs (Fig. 1C), (iv) lysin bound to this material with high (nanomole) affinity (Fig. 3A), (v) the species selectivity of the interaction of lysin with the isolated high molecular mass material (Fig. 3A) agreed with the species selectivity of lysin in dissolving intact VEs (Fig. 3D).
SDS/polyacrylamide gels of VERL (Fig. 2 A and C) showed it was composed of 1–3 silver-staining bands migrating between titin (ref. 21; 2,800 kDa) and nebulin (770 kDa). One-dimensional peptide maps showed these components derived from the same protein backbone (Fig. 2B). The differential mobility could result from proteolysis, differential glycosylation, or charge differences. The long, rod-like appearance of VERL by electron microscopy (Fig. 2D) is consistent with a large molecular mass.
In addition to showing that VERL is a distinct VE component, its amino acid composition shows it to be acidic (22% Glu and Asp). Ignoring charge due to glycosylation, the net negative charge (Glu + Asp − Arg + Lys) of VERL for both species would be −13. This compares to the net positive charge for Hc lysin of +16 and of Hr lysin of +13. These charge differences support the hypothesis that interaction of lysin and VERL could be by hydrogen bond disruption; electrostatic interaction is excluded as a possibility, because the activity of lysin to dissolve VEs is unaffected by 3 M NaCl (unpublished work). The possibility that both hydrogen bond displacement and hydrophobic interaction by the hydrophobic patch of lysin are involved in lysin’s mechanism of action has been discussed (19). Recognition between VERL and lysin could involve the carbohydrate moieties of VERL. Peptide-N-glycosidase-F digestion of VERL does not alter its mobility on SDS/2.5% polyacrylamide gels. Amino acid analysis (Table 1) showing that 20% of the residues are Thr and Ser indicate the potential for extensive O-linked glycosylation.
Stoichiometric binding assays showed that each molecule of VERL binds more than 100 lysin monomers. Furthermore, the binding shows positive cooperativity, indicating that binding is complex and that more than one class of lysin binding sites exists. In the heterospecific mixture of Hc VERL and Hr lysin, more lysin is required to initiate binding. However, once binding begins, all curves ascend in roughly parallel fashion, suggesting that species-specific recognition is in the initial interaction of lysin with VERL. The other VE glycoproteins of 30–50 kDa, although not binding lysin, could be involved in mediating species selectivity based on their interaction with VERL. However, lysin binding data, using the total pH 2.8-soluble VE material, showed binding curves similar to those with isolated VERL (Fig. 3A), suggesting these lower molecular weight VE components do not govern species-selective recognition.
The Apparent Giant Size of VERL.
Because VERL remains of large size when heated in SDS and mercaptoethanol, it is a covalently bonded structure. Few biological molecules are known of this size, muscle titin (21) being one such protein. However, although VERL is a giant molecule, it is unknown if it is translated from a single species of mRNA. At least three hypotheses could account for the giant size of VERL. (i) VERL, like titin, could be translated from a single mRNA. (ii) VERL precursor proteins could be translated from one species of mRNA and then linked together by, for example, a transglutaminase reaction. (iii) VERL could be created by the covalent linkage of several nonidentical translation products. Molecular cloning of VERL will distinguish among these hypotheses.
Acknowledgments
We thank Drs. C. D. Stout, R. A. Cardullo, R. F. Doolittle, P. G. A. Fortes, J. D. Bleil, D. P. Millar, J. D. Calkins, J. R. Schulz, and J. M. Swanson for helpful discussions and R. R. McConnaughey for collecting abalone. The work was supported by National Institutes of Health Grant HD12986.
ABBREVIATIONS
- VE
vitelline envelope
- VERL
VE receptor for lysin
- Hc
abalone species Haliotis corrugata
- Hr
abalone species Haliotis rufescens
- HSW
millipore-filtered seawater buffered with 10 mM Hepes (pH 7.8)
- ZP
zona pellucida
References
- 1.Metz E C, Palumbi S R. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 2.Vacquier V D, Swanson W J, Hellberg M E. Dev Growth Differ. 1995;37:1–10. doi: 10.1046/j.1440-169X.1995.00001.x. [DOI] [PubMed] [Google Scholar]
- 3.Vacquier V D, Lee Y-H. Zygote. 1993;1:181–196. doi: 10.1017/s0967199400001465. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y-H, Ota T, Vacquier V D. Mol Biol Evol. 1995;12:231–238. doi: 10.1093/oxfordjournals.molbev.a040200. [DOI] [PubMed] [Google Scholar]
- 5.Palumbi S R. Trends Ecol Evol. 1992;7:114–118. doi: 10.1016/0169-5347(92)90144-Z. [DOI] [PubMed] [Google Scholar]
- 6.Swanson W J, Vacquier V D. Proc Natl Acad Sci USA. 1995;92:4957–4961. doi: 10.1073/pnas.92.11.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis C A, Talbot C F, Vacquier V D. Dev Biol. 1982;92:227–239. doi: 10.1016/0012-1606(82)90167-1. [DOI] [PubMed] [Google Scholar]
- 8.Mozingo N M, Vacquier V D, Chandler D E. Mol Reprod Dev. 1995;41:493–502. doi: 10.1002/mrd.1080410412. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman P M, Liu C, Litscher E S. J Cell Sci. 1996;109:2001–2004. doi: 10.1242/jcs.109.8.2001. [DOI] [PubMed] [Google Scholar]
- 10.Myles D G. Dev Biol. 1993;158:35–45. doi: 10.1006/dbio.1993.1166. [DOI] [PubMed] [Google Scholar]
- 11.Richardson R T, O’Rand M G. J Biol Chem. 1996;271:24069–24074. [PubMed] [Google Scholar]
- 12.Bookbinder L H, Cheng A, Bleil J D. Science. 1995;269:86–89. doi: 10.1126/science.7604284. [DOI] [PubMed] [Google Scholar]
- 13.Ward C R, Kopf G S. Dev Biol. 1993;158:9–34. doi: 10.1006/dbio.1993.1165. [DOI] [PubMed] [Google Scholar]
- 14.Foltz K R. Int Rev Cytol. 1995;163:249–303. doi: 10.1016/s0074-7696(08)62212-3. [DOI] [PubMed] [Google Scholar]
- 15.Ohlendieck K, Lennarz W J. Trends Biochem Sci. 1995;20:29–33. doi: 10.1016/s0968-0004(00)88947-1. [DOI] [PubMed] [Google Scholar]
- 16.Moy G W, Mendoza L M, Schulz J R, Swanson W J, Glabe C G, Vacquier V D. J Cell Biol. 1996;133:809–817. doi: 10.1083/jcb.133.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw A, McRee D E, Vacquier V D, Stout C D. Science. 1993;262:1864–1867. doi: 10.1126/science.8266073. [DOI] [PubMed] [Google Scholar]
- 18.Shaw A, Lee Y-H, Stout C D, Vacquier V D. Semin Dev Biol. 1994;5:209–215. [Google Scholar]
- 19.Shaw A, Fortes P G A, Stout C D, Vacquier V D. J Cell Biol. 1995;130:1117–1125. doi: 10.1083/jcb.130.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Granzier H L M, Wang K. Electrophoresis. 1993;14:56–64. doi: 10.1002/elps.1150140110. [DOI] [PubMed] [Google Scholar]
- 22.Morrissey J H. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 23.Merril C R. Methods Enzymol. 1990;182:477–488. doi: 10.1016/0076-6879(90)82038-4. [DOI] [PubMed] [Google Scholar]
- 24.Jameson D M, Sawyer W H. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 25.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Anal Chem. 1956;28:350–356. [Google Scholar]
- 26.Correa L M, Carroll E J. Dev Growth Differ. 1997;39:69–85. doi: 10.1046/j.1440-169x.1997.00008.x. [DOI] [PubMed] [Google Scholar]
- 27.Focarelli R, Rosati F. Dev Biol. 1995;171:606–614. doi: 10.1006/dbio.1995.1308. [DOI] [PubMed] [Google Scholar]
- 28.Litscher E S, Honegger T G. Dev Biol. 1991;148:536–551. doi: 10.1016/0012-1606(91)90272-5. [DOI] [PubMed] [Google Scholar]
- 29.Downey J C, Lambert C C. Mol Reprod Dev. 1994;38:453–458. doi: 10.1002/mrd.1080380413. [DOI] [PubMed] [Google Scholar]
- 30.Godknecht A J, Honegger T G. Dev Growth Differ. 1995;37:183–189. doi: 10.1046/j.1440-169X.1995.t01-1-00007.x. [DOI] [PubMed] [Google Scholar]
- 31.Omata S, Katagiri C. Dev Growth Differ. 1996;38:663–672. doi: 10.1046/j.1440-169X.1996.t01-5-00010.x. [DOI] [PubMed] [Google Scholar]
- 32.Tian J, Gong H, Thomsen G H, Lennarz W J. J Cell Biol. 1997;136:1099–1108. doi: 10.1083/jcb.136.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litscher E S, Wassarman P M. Zygote. 1996;4:229–236. doi: 10.1017/s0967199400003142. [DOI] [PubMed] [Google Scholar]
- 34.Litscher E S, Wassarman P M. Biochemistry. 1996;35:3980–3985. doi: 10.1021/bi952722m. [DOI] [PubMed] [Google Scholar]
- 35.Thaler C D, Cardullo R A. J Biol Chem. 1996;271:23289–23297. doi: 10.1074/jbc.271.38.23289. [DOI] [PubMed] [Google Scholar]