Abstract
Transcription factors of the AML (core binding factor-α/polyoma enhancer binding protein 2) class are key transactivators of tissue-specific genes of the hematopoietic and bone lineages. Alternative splicing of the AML-1 gene results in two major AML variants, AML-1 and AML-1B. We show here that the transcriptionally active AML-1B binds to the nuclear matrix, and the inactive AML-1 does not. The association of AML-1B with the nuclear matrix is independent of DNA binding and requires a nuclear matrix targeting signal (NMTS), a 31 amino acid segment near the C terminus that is distinct from nuclear localization signals. A similar NMTS is present in AML-2 and the bone-related AML-3 transcription factors. Fusion of the AML-1B NMTS to the heterologous GAL4-(1–147) protein directs GAL4 to the nuclear matrix. Thus, the NMTS is necessary and sufficient to target the transcriptionally active AML-1B to the nuclear matrix. The loss of the C-terminal domain of AML-1B is a frequent consequence of the leukemia-related t(8;21) and t(3;21) translocations. Our results suggest this loss may be functionally linked to the modified interrelationships between nuclear structure and gene expression characteristic of cancer cells.
The gene encoding transcription factor AML-1 [core binding factor α (CBF-α)/polyoma enhancer binding protein 2 (PEBP2)] is frequently the target of multiple chromosomal translocations in lymphoid and myeloid leukemias (1–10). Human AML-1 recognizes the core motif 5′-TGYGGT (Y = C or T) and heterodimerizes with the non-DNA-binding partner CBF-β (1, 11–14). The AML-1 class of proteins includes AML-2 and AML-3, and also the murine homologs PEBP 2αA and BEBP 2αB, which share a runt homology DNA-binding domain (rhd) first documented in the Drosophila runt gene (12–18). The human AML-1 gene comprises at least eight different exons and multiple polyadenylylation sites (19) and encodes multiple alternatively spliced proteins. The largest of these is AML-1B (480 aa), a transcriptional activator (14, 17). The shorter AML-1 protein (250 aa) does not activate transcription (13, 14). The major distinction between inactive AML-1 and transcriptionally active AML-1B is use of the alternative C-terminal exons 7B and 8 that are absent in AML-1. These exons are separated from the rhd by many leukemia-associated chromosomal translocations.
AML-related factors are expressed in a variety of tissues including lymphoid, myeloid, and osteoblast lineages (1, 17, 18, 20–22), where they are key components of mechanisms mediating tissue-specific transcription (22–29). Recent studies have shown that the nuclear matrix protein NMP-2, specific to osseous cells, is an AML-related protein (21) and that AML transcription factors activate the bone-specific osteocalcin promoter through an NMP-2 binding site (22).
Regulation of gene expression is linked to organization of nuclear structure, a principal component of which is the filamentous ribonucleoprotein network known as the nuclear matrix. The nuclear matrix is involved in gene localization and in the concentration and subnuclear localization of regulatory factors (30–35). A direct demonstration of the specificity of transcription factor/nuclear matrix interactions would be a significant advance for understanding the role of nuclear structure in regulating gene expression. Here, we show that transcriptionally active AML-1B associates with the nuclear matrix but inactive AML-1 does not. Consequently, a fundamental question is how active AML transcription factors become associated with the nuclear matrix. We show that sequences required for targeting AML-1B to the nuclear matrix reside in a 31-aa segment [nuclear matix targeting signal (NMTS), amino acids 351–381] within the C-terminal domain. The NMTS is physically distinct from the nuclear localization signal, functions autonomously, and is closely associated with the transactivation domain.
METHODS
Transient Transfections.
Expression vectors were based on pcDNA/ampI and contain the cytomegalovirus (CMV) promoter and segments of the human AML-1B cDNA linked to the hemagglutinin (HA)-tag fused in frame with amino acid 27 of AML-1B (CMV/HA/AML-1B constructs). DEAE-dextran-mediated transfection was performed with 15–20 μg of plasmid DNA mixed with 1.5 ml of F12 serum-free medium containing 0.2 mg/ml DEAE-dextran and 0.05 mg/ml chloroquine. Aliquots of 0.5 ml of this mixture were added to each well of a six-well plate and incubated for 1.5–2.0 hr at 37°C. Cells were subject to a 15% glycerol shock for 2 min, washed with PBS, refed, and harvested at 36–40 hr following transfection.
Cellular Fractionation.
Nuclear matrix fractions were prepared by sequential extraction (36) with CSK buffer, RSB buffer, and digestion buffer followed by 0.25 M ammonium sulfate extraction. Fractions for the salt-resistant nuclear retention assay were prepared by sequential incubations in IsoHi buffer (10 mM Tris⋅Cl, pH 8.4/140 mM NaCl/1.5 mM MgCl2/0.5% Nonidet P-40) and high salt buffer (20 mM Hepes, pH 7.9/1 M NaCl/0.2 mM EDTA/20% glycerol/1.5 mM MgCl2/0.1 mM EGTA/1.2 mM phenylmethylsulfonyl fluoride). Western blot analysis was performed after electrophoresis in SDS/10% or 12% polyacrylamide gels, and proteins were detected with AML or HA antibodies (1:3,000 dilution) using enhanced chemiluminescence (Amersham).
In Situ Nuclear Matrix Isolation and Indirect Immunofluorescence Analysis.
Cells on coverslips were washed in PBS and extracted twice in CSK buffer. DNase digestion was performed twice in digestion buffer (CSK buffer but with 100 μg/ml DNase I and 50 mM NaCl) followed by extraction in digestion buffer containing 0.25 M (NH4)2SO4. The coverslips were fixed in 4% formaldehyde in PBS. Whole cell samples were fixed directly after the PBS wash, followed by permeabilization with 0.25% Triton X-100 in PBS. The primary antibody was incubated for 1–1.5 hr at 37°C. The primary antibodies were anti-HA (1:1,500 dilution, 12CA5 mouse monoclonal, a gift from M. Czech, University of Massachusetts Medical Center, Worcester), anti-NuMa (1:200 dilution rabbit polyclonal, a gift from Matritech, Newton, MA), anti-AML-1 (1:200 dilution, rabbit polyclonal antibody against the N-terminal peptide of AML-1). The secondary antibody was incubated for 1 hr at 37°C and was either a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (1:400, Jackson ImmunoResearch) or a Texas red-conjugated donkey anti-mouse antibody (1:400, Jackson ImmunoResearch). DNA content was evaluated by 4′,6-diamidino-2-phenylindole (DAPI) staining (5 μg/ml DAPI in PBS containing BSA and 0.05% Triton X-100). Cells were mounted in Vectashield H-1000.
RESULTS
The Transcription Factor AML-1B Is Associated with the Nuclear Matrix.
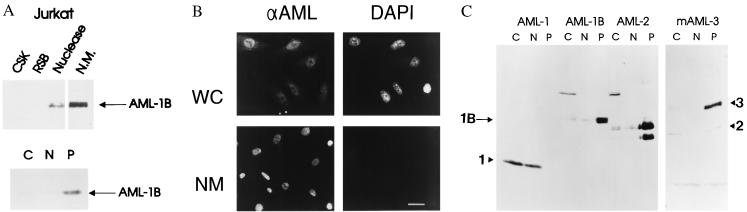
We initially investigated subcellular partitioning of AML-1B in Jurkat T cells using two different extraction protocols. Nuclear matrix/intermediate filament preparations were obtained from Jurkat cells. Western blot analysis shows that most of AML-1B is associated with the nuclear matrix fraction (Fig. 1A Upper), which is also positive for the nuclear matrix specific marker lamin B (data not shown). Thus, AML factors are present in the nuclear matrix in cells of both bone (21, 35) and hematopoietic lineages. Like many other NMPs, AML-1B is not extracted from nuclei by high salt (14) (Fig. 1A Lower). We also visualized the cellular distribution of AML proteins by immunofluorescence analysis in ROS 17/2.8 osteosarcoma cells. Biochemical analysis previously showed that ROS 17/2.8 cells contain AML-related transcription factors in their nuclear matrix (21). Fig. 1B shows that endogenous AML-related factors are retained throughout the nuclear matrix preparations from which all chromatin detectable by DAPI staining has been removed. This finding supports the specific association of AML-related factors with the nuclear matrix.
Figure 1.
AML-1B is associated with the nuclear matrix. (A) AML-1B is present in the biochemical nuclear matrix and the salt-resistant insoluble nuclear fraction. Biochemical fractionation of Jurkat cells using the nuclear matrix protocol (Upper) yields cytoplasmic (CSK and RSB), chromatin (Nuclease), and nuclear matrix (N.M.) fractions. Nuclear extraction of Jurkat cells (Lower) results in cytoplasmic (C), salt-extractable nuclear (N), and high salt-resistant nuclear “pellet” (P) fractions. Protein representing equal cell numbers was loaded in each lane. Western blot analysis was performed using an affinity-purified rabbit polyclonal antibody against the 17 aa at the N terminus of AML-1. (B) Immunofluorescence localization of AML-related proteins in ROS 17/2.8 cells. Whole cell (WC) and in situ nuclear matrix (NM) preparations were analyzed for the presence of AML-related proteins by indirect immunofluorescence with the AML-1 anti-peptide antibody (Left). DNA content was assessed by DAPI staining (Right). (C) Salt-resistant nuclear retention is a shared property among AML proteins. AML-1, AML-1B, AML-2, and AML-3 cDNAs were expressed in Cos-7 cells transfected by the DEAE-dextran procedure (14). Subcellular fractions (as described in A) were subjected to Western blot analysis with AML-specific antibodies. AML-1 (250 aa) is a splice variant of AML-1B (480 aa) that lacks the C-terminal extension that is conserved among AML-1B, AML-2, and AML-3.
We analyzed the subcellular partitioning of several distinct AML proteins to determine whether these might also be associated with the nuclear matrix. AML-1B, AML-2, and AML-3 are encoded by three different genes, but each possesses a similar C-terminal extension that is absent from AML-1, the 33-kDa splice variant of the AML-1 gene. We expressed these proteins in Cos-7 cells. The results show clearly that AML-1B, AML-2, and AML-3 remain in the high-salt-resistant fraction, while AML-1 is extracted into both cytoplasmic and nuclear fractions and is absent from the salt-resistant fraction (Fig. 1C). Hence, it appears that molecular differences between AML-1 and AML-1B result in specific differences in their partitioning between subnuclear fractions. The observation that AML-1B, AML-2, and AML-3 all localize to the nuclear matrix-containing fraction suggests that association with the nuclear matrix is a property shared among these three AML family members.
Nuclear Matrix Targeting of AML-1B Is Independent of DNA Binding.
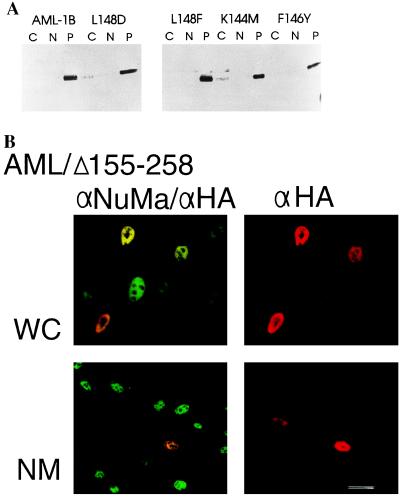
The rhd is the most strongly conserved protein segment among the AML class of transcription factors, and it mediates both DNA binding and protein–protein interactions with the AML partner CBF-β. To assess whether these two functions are required for salt-resistant nuclear retention, we analyzed subnuclear partitioning of AML-1B proteins containing single amino acid substitutions in the rhd (37). One mutation affects DNA binding but not CBF-β interaction (K144M), others eliminate both activities (L148D, F146Y), while another does not affect wild-type function (L148F). Each of these mutant proteins remained in the salt-resistant insoluble nuclear fraction (Fig. 2A), showing that neither DNA binding nor CBF-β interaction is required for association with the nuclear matrix.
Figure 2.
Nuclear matrix association of AML-1B is independent of DNA binding and CBF-β interaction. (A) Subcellular partitioning of AML-1B/rhd substitution mutants that affect DNA binding and/or CBF-β interactions (37). Nuclear extraction results in cytoplasmic (C), salt-extractable nuclear (N), and salt-resistant nuclear “pellet” (P) fractions. (B) In situ immunofluorescence analysis of the HA-tagged AML-1B/Δ155–258 protein expressed in ROS 17/2.8 cells; this protein lacks a portion of the rhd and is defective for DNA binding. Immunostaining with a NuMa antibody (αNuMa) and/or an HA antibody (αHA) for detection of HA/AML is observed in both whole cell (WC) and in situ nuclear matrix preparations (NM). (Bar = 10 μm.)
Further evidence for independence of nuclear matrix attachment and DNA binding was afforded by an HA-tagged mutant of AML-1B with an internal deletion of amino acids 155–258 (AMLΔ155–258). This mutation removes the distal portion of the rhd encompassing critical motifs involved in DNA binding (38) and an auxiliary determinant for nuclear localization (39). This mutant protein does not bind DNA but nevertheless resists extraction from the nuclear matrix (Fig. 2B). Hence, DNA binding is not necessary for nuclear matrix association in situ.
The C-Terminal Domain of AML-1B Is Required for Targeting to the Nuclear Matrix.
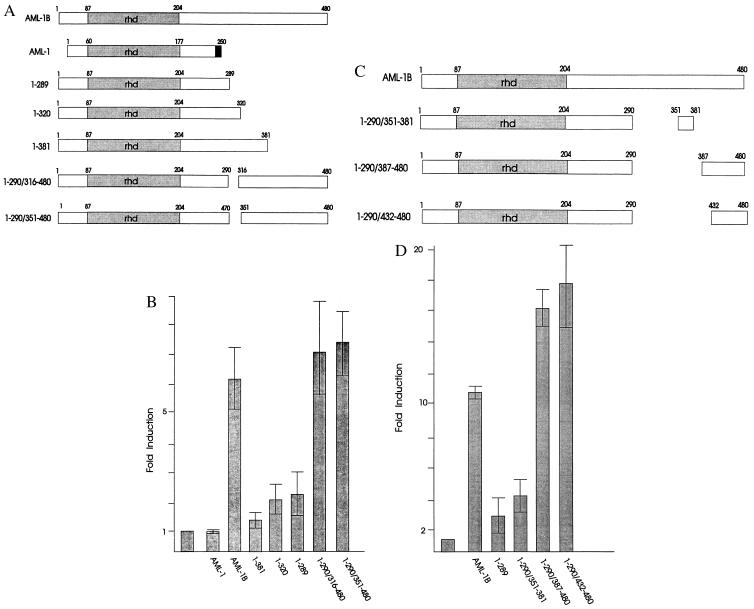
AML-1B is associated with the nuclear matrix, while AML-1 is not. An obvious difference between the two proteins is the C-terminal extension of AML-1B that is absent in AML-1 (see Fig. 4A). This protein segment contains a strong transcriptional activation domain (15). We established the importance of the C-terminal domain in linking AML to the nuclear matrix by constructing a version of AML-1B (AML1–289) that is truncated beyond amino acid 289 and resembles the AML-1 (33-kDa) splice variant (see Fig. 4). We found a marked difference in the partitioning of the two proteins in biochemical fractionation: AML-1B remains entirely in the nuclear matrix fraction, while AML1–289 is not bound to the matrix but elutes completely with the chromatin fraction (data not shown).
Figure 4.
Delineation of the transactivation domain in the C terminus of AML-1B. (A and C) Diagrams of C-terminal and internal deletion mutants of AML-1B. Dark shading designates sequences unique to AML-1. (B and D) Transactivation analysis of AML-1B deletion mutants using the AML-1B-responsive reporter construct TCRβ-CAT. The bar graph shows the fold induction of transcription with each AML protein as the average of three separate experiments. (Bar = SD.) C33A cells were transfected by the calcium phosphate method (14) using 1 μg of TCRβ-CAT (25), pCMV5, or pCMV5 vectors expressing AML-1 or the AML-1B deletion mutants, and 5 μg of pRSV-long terminal repeat/SEAP expressing a secreted alkaline phosphatase gene (SEAP) (40). pBlueScript was added to bring each transfection to 25 μg of total DNA. Promoter activity was normalized for transfection efficiency by using SEAP.
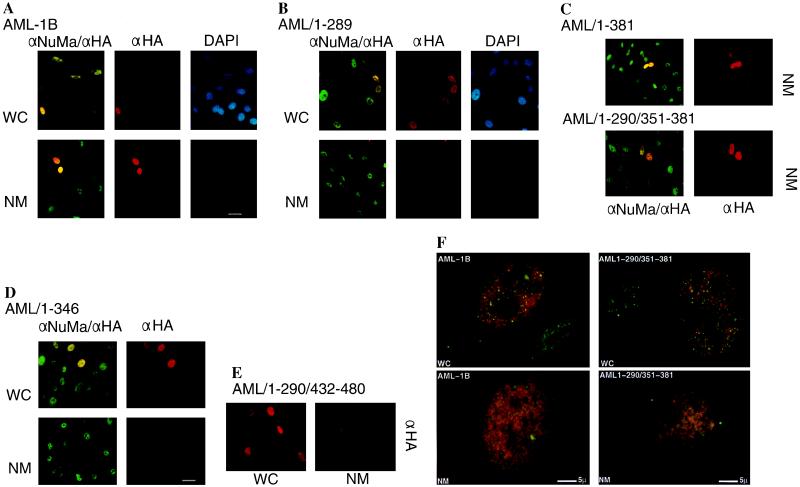
We also carried out in situ immunofluorescence analysis of whole cells and nuclear matrices in transfected ROS 17/2.8 cells expressing HA-tagged AML proteins (Fig. 3). Visualization of the nuclear matrix was achieved by immunostaining with an antibody that detects the nuclear matrix protein NuMa (Fig. 3). Both AML-1B and AML1–289 proteins show a similar broad distribution throughout the nucleus. However, both biochemical and in situ assays show that AML1–289 is not present in the nuclear matrix, while AML-1B is nuclear matrix associated. We conclude that the C terminus of AML-1B (amino acids 290–480) contains a segment essential for association with the nuclear matrix.
Figure 3.
Delineation of the AML-1B NMTS by in situ immunofluorescence analysis. Wild-type HA/AML-1B (A), HA/AML1–289 (B), HA/AML1–381 and HA/AML1–290/351–381 (C), HA/AML1–346 (D), and HA/AML1–290/432–480 (E) were transiently transfected into ROS 17/2.8 cells and analyzed in whole cell (WC) or in situ nuclear matrix (NM) preparations. Preparations were visualized with a fluorescein isothiocyanate-conjugated secondary antibody detecting NuMa (αNuMa; green) and a Texas red-conjugated antibody detecting the HA epitope (αHA; red). DNA was visualized by DAPI staining (DAPI; blue). (Bar = 10 μm.) Regions of colocalization appear yellow. However, high-resolution, computer-enhanced images reveal that the staining patterns for NuMa and AML-1B are dispersed and punctate but nonoverlapping (F). Constructs lacking the NMTS (AML1–346 and AML1–290/432–480) localize in the nucleus but are not retained in the nuclear matrix.
The NMTS of AML-1B Resides Between Amino Acids 351 and 381.
To establish the specific domains in the C-terminal region of AML-1B required for binding to the nuclear matrix and for transactivation, we constructed a series of HA-tagged C-terminal deletion mutants, as well as mutants with deletions in internal domains of AML-1B (see Fig. 4). We determined the subcellular location of these AML-1B mutants by indirect immunofluorescence labeling of nuclear matrix preparations (Fig. 3). The data show that AML1–381 and AML1–290/351–381, but not AML1–346 and AML1–290/432–480, associate with the nuclear matrix of ROS 17/2.8 cells (Fig. 3). Western blot analysis of subcellular fractions corroborates these findings (data not shown). We conclude that amino acids between 351 and 381 contain a unique NMTS that is necessary for directing the rhd-containing portion of AML-1B (amino acids 1–289) to the nuclear matrix.
The Transactivation Domain of AML-1B (Amino Acids 432–480) and the NMTS (Amino Acids 351–381) Are Both Encoded by Exon 8.
The C terminus of AML-1B (amino acids 290–480) contains, in addition to the NMTS, a potent transactivation (TA) domain. The absence of this region in AML1–289 correlates with dramatically reduced transactivation potential (Fig. 4). We mapped the specific domain in the C-terminal region of AML-1B required for transactivation by using a series of C-terminal and internal deletion mutants (Fig. 4). Deletion of C-terminal residues 382–480 (AML1–381) abolishes transactivation (Fig. 4). Internal deletions of either 291 to 315 (AML1–290/316–480) or 291 to 350 (AML1–290/351–480) do not significantly reduce the transactivation potential of AML-1B (Fig. 4). We also observed that AML1–290/387–480 and AML1–290/432–480 proteins are maximally active, whereas AML1–290/351–381 is not (Fig. 4). These results clearly show that the main TA domain of AML-1B is found at the C terminus between amino acids 432 and 480 and is separable in vitro from the NMTS. However, both the TA domain (amino acids 432–480) and the NMTS (amino acids 351–381) are encoded by the same exon (exon 8). Thus, although the TA and NMTS domains can be separated experimentally, the transactivation and nuclear matrix targeting functions are not dissociated in vivo.
The NMTS Is Sufficient to Target a Heterologous Nuclear Protein to the Nuclear Matrix.
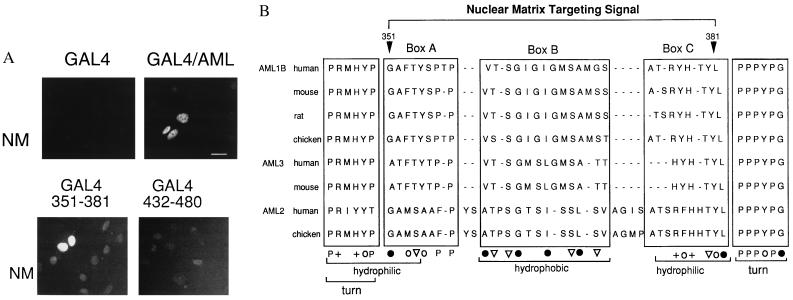
If the C-terminal segment of AML-1B contains a targeting signal and represents an autonomous protein domain, it should direct a heterologous protein to the nuclear matrix. We compared binding to the nuclear matrix in cells expressing GAL4-(1–147) protein (41) alone or chimeric proteins with GAL4-(1–147) fused to the C terminus of AML-1B (amino acids 171–480), to the NMTS (amino acids 351–381), or to the TA domain (amino acids 432–480). Nuclear matrices were labeled for immunofluorescence with an antibody to GAL4. Fig. 5 shows that both the GAL4/AML-1B C-terminal fusion protein and the GAL-4/AML351–381 fusion protein are bound to the nuclear matrix. In contrast, although the GAL4-(1–147) protein and the GAL-4/AML432–480 fusion protein are transported into the nucleus, these proteins do not bind to the nuclear matrix. We conclude that the NMTS of AML-1B (amino acids 351–381) (Fig. 5B) is necessary and sufficient for targeting this protein to the nuclear matrix.
Figure 5.
The NMTS is sufficient to direct a heterologous nuclear protein to the nuclear matrix. (A) In situ immunofluorescence analysis of nuclear matrices prepared from ROS 17/2.8 cells transfected with expression vectors for GAL4-(1–147), GAL4-(1–147)/AML171–480, GAL4/AML351–381, and GAL4/AML432–480 as indicated. Fusion proteins were visualized using a GAL4-specific primary antibody and a fluorescein isothiocyanate-conjugated secondary antibody. (Bar = 10 μm.) (B) Comparison of the amino acid sequence of the AML-1B NMTS with sequences of other members of the AML family, including the bone-related AML-3 factor (62, 63). Three motifs (boxes A, B, and C) show conservation of amino acids with either aliphatic (•), aromatic (○), aliphatic hydroxyl (▿), or basic (+) side chains. The brackets denote segments with a propensity to form turns or a preponderance of hydrophobic or hydrophilic residues, based on the Kyte–Doolittle hydropathy plot using Genetics Computer Group (Madison, WI) software. The NMTS is devoid of negatively charged amino acids; 20 of 31 amino acids are G, A, Y, T, or S.
DISCUSSION
Nuclear Architecture and Transcription Factor Targeting.
The nuclear matrix may contribute to control of gene expression by localizing and/or concentrating transcription factors. We have defined a 31-amino acid segment (NMTS) that mediates association of transcriptionally active AML-1B with the nuclear matrix, and we have shown a close physical linkage of this NMTS to the transactivation domain. Furthermore, the transcriptionally inactive AML-1 splice variant lacks the NMTS. Our results provide insight into mechanisms by which gene-regulatory factors are targeted to the nuclear matrix. The existence of a nuclear matrix targeting module that functions independent of the AML-1B DNA-binding domain provides evidence for the specificity of these factor/nuclear matrix interactions. Specific targeting argues against indiscriminate attachment of such proteins to the nuclear matrix during subcellular fractionation.
The functional complexity of nuclear organization is reflected by the multiplicity of specialized nuclear substructures contributing to DNA replication and/or gene expression, including the nucleolus, RNA polymerase II transcription and processing domains, coiled bodies, promyelocytic-leukemia factor (PML) domains, and Barr bodies (42–50). The nuclear matrix, the anastomosing network of nuclear filaments, may provide the underlying structure that supports nuclear compartmentalization, and is itself a specialized nuclear structure. The nuclear matrix mediates long-range control of gene transcription, and several DNA-binding proteins involved in gene/matrix interactions have been identified, including SATB-1 (51), ARBP (52, 53), lamin B (54, 55), NMP-1 (56), and NMP-2 (21, 35). The nuclear matrix is a developmentally modulated structure as reflected by its protein composition that is specific to tissue type and differentiation stage (reviewed in refs. 30 and 31). One basic question is how proteins traffic selectively to such specialized domains within the nucleus to become components of functional complexes.
Previous data and the current study show that at least two trafficking signals are required for subnuclear targeting of AML transcription factors; the first supports nuclear import (nuclear localization signal) and a second mediates association with the nuclear matrix (NMTS). Lu et al. (39) observed that both the N-terminal (amino acids 94–221) and C-terminal (amino acids 221–513) regions of the murine AML-1 homolog contribute to nuclear import. Our data show that amino acids between 351 and 381 of human AML-1B confer nuclear matrix association (NMTS). The NMTS and the transactivation domain of AML-1B are both encoded by exon 8. Although the NMTS and transactivation domains can be separated by in vitro manipulation, in vivo splicing of exon 8 will always result in AML proteins that contain both functional domains. The multiplicity of determinants for nuclear localization and alternative splicing of AML mRNAs may provide the requisite complexity to support targeting to specific sites within the nucleus in response to diverse biological conditions. Furthermore, because gene regulation by AML-1B involves contributions by other factors—such as CBF-β (11, 12), ets-1 (57), and C/EBP (58)—AML-1B may facilitate recruitment of these factors to the nuclear matrix.
Implications of Chromosomal Translocation for Transcription Factor Targeting in Leukemic Cells.
AML-1 is one of the most frequently affected genes in human leukemia. Mapping of the nuclear matrix attachment site to exon 8 reveals that this domain is not present in the t(8;21) fusion protein (AML-1/ETO), but is replaced by sequences from the MTG8 gene (59, 60). Thus, intranuclear targeting of the AML transcription factor may be abrogated because of chromosomal translocations in leukemic cells. Fidelity of transcriptional control may involve the localization of gene-regulatory proteins to the correct subnuclear region. For example, PML bodies are nuclear structures that are associated with the nuclear matrix and modified in promyelocytic leukemia cells (30, 49, 61). In normal cells the PML protein resides in discrete PML bodies. However, in leukemic cells the PML protein is genetically rearranged and dispersed throughout the nucleus. We suggest that perturbations in subnuclear location and/or nuclear matrix association of proteins may be related to modifications in gene expression that are linked to leukemias. These perturbations may also effect the alterations in nuclear organization that are the hallmarks of cancer cells.
Acknowledgments
We thank Liz Allison for helpful comments, Elizabeth Bronstein for editorial assistance, and John McNeil for high-resolution image analysis. These studies were supported by National Institutes of Health Grants AR42262 and CA64140 and by the Our Danny Cancer Fund.
ABBREVIATIONS
- NMP
nuclear matrix protein
- NMTS
nuclear matrix targeting signal
- PML
promyelocytic-leukemia factor
- DAPI
4′,6-diamidino-2-phenylindole
- HA
hemagglutinin
- TA
transactivation
- rhd
runt homology domain
References
- 1.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nisson P E, Watkins P C, Sacchi N. Cancer Genet Cytogenet. 1992;63:81–88. doi: 10.1016/0165-4608(92)90384-k. [DOI] [PubMed] [Google Scholar]
- 3.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 4.Downing J R, Head D R, Curcio-Brint A M, Hulshof M G, Motroni T A, Raimondi S C, Carroll A J, Drabkin H A, Willman C, Theil K S, Civin C I, Erickson P. Blood. 1993;81:2860–2865. [PubMed] [Google Scholar]
- 5.Mitani K, Ogawa S, Tanaka T, Miyoshi H, Kurokawa M, Mano H, Yazaki Y, Ohki M, Hirai H. EMBO J. 1994;13:504–510. doi: 10.1002/j.1460-2075.1994.tb06288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubin C M, Larson R A, Anastasi J, Winter J N, Thangavelu M, Vardiman J W, Rowley J D, LeBeau M M. Blood. 1990;76:2594–2598. [PubMed] [Google Scholar]
- 7.Nucifora G, Birn D J, Espinosa R I, Erickson P, LeBeau M M, Roulston D, McKeithan T W, Drabkin H, Rowley J D. Blood. 1993;81:2728–2734. [PubMed] [Google Scholar]
- 8.Romana S P, Mauchauffe M, Le Coniat M, Chumakov I, Le Paslier D, Berger R, Bernard O A. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- 9.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romana S P, Poirel H, Leconiat M, Flexor M-A, Mauchauffe M, Jonveaux P, Macintyre E A, Berger R, Bernard O A. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- 11.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyers S, Downing J R, Hiebert S W. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers S, Lenny N, Hiebert S W. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 16.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 18.Meyers S, Lenny N, Sun W-H, Hiebert S W. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 19.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Mol Cell Biol. 1995;15:1662–1670. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 22.Banerjee C, Hiebert S W, Stein J L, Lian J B, Stein G S. Proc Natl Acad Sci USA. 1996;93:4968–4973. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuchprayoon I, Meyers S, Scott L M, Suzow J, Hiebert S W, Friedman A D. Mol Cell Biol. 1994;14:5558–5568. doi: 10.1128/mcb.14.8.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho I-C, Yang L-H, Morle G, Leiden J M. Proc Natl Acad Sci USA. 1989;86:6714–6718. doi: 10.1073/pnas.86.17.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottschalk L R, Leiden J M. Mol Cell Biol. 1990;10:5486–5495. doi: 10.1128/mcb.10.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez-Munain C, Krangel M S. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 28.Takahashi A, Satake M, Yamaguchi-Iwai Y, Bae S C, Lu J, Maruyama M, Zhang Y W, Oka H, Arai N, Arai K-I, Ito Y. Blood. 1995;86:607–616. [PubMed] [Google Scholar]
- 29.Nimer S, Zhang J, Avraham H, Miyazaki Y. Blood. 1996;88:66–74. [PubMed] [Google Scholar]
- 30.Nickerson J A, Blencowe B J, Penman S. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 31.Stein G S, van Wijnen A J, Stein J L, Lian J B, Bidwell J P, Montecino M. J Cell Biochem. 1994;55:4–15. doi: 10.1002/jcb.240550103. [DOI] [PubMed] [Google Scholar]
- 32.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mancini M A, Shan B, Nickerson J A, Penman S, Lee W-H. Proc Natl Acad Sci USA. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Wijnen A J, Bidwell J P, Fey E G, Penman S, Lian J B, Stein J L, Stein G S. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- 35.Bidwell J P, van Wijnen A J, Fey E G, Dworetzky S, Penman S, Stein J L, Lian J B, Stein G S. Proc Natl Acad Sci USA. 1993;90:3162–3166. doi: 10.1073/pnas.90.8.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fey E G, Wan K M, Penman S. J Cell Biol. 1984;98:1973–1984. doi: 10.1083/jcb.98.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenny N, Meyers S, Hiebert S. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 38.Kurokawa M, Tanaka T, Tanaka K, Hirano N, Ogawa S, Mitani K, Yazaki Y, Hirai H. J Biol Chem. 1996;271:16870–16876. doi: 10.1074/jbc.271.28.16870. [DOI] [PubMed] [Google Scholar]
- 39.Lu J, Maruyama M, Satake M, Bae S C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger J, Hauber J, Hauber R, Geiger R, Cullen B R. Gene. 1988;66:1–10. doi: 10.1016/0378-1119(88)90219-3. [DOI] [PubMed] [Google Scholar]
- 41.Silver P A, Keegan L P, Ptashne M. Proc Natl Acad Sci USA. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter K C, Bowman D, Carrington W, Fogarty K, McNeil J A, Fay F S, Lawrence J B. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 43.Xing Y, Johnson C V, Dobner P R, Lawrence J B. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 44.Fakan S. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 45.Scheer U, Thiry M, Goessens G. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- 46.Spector D L. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 47.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 48.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 50.Matera A G, Ward D C. J Cell Biol. 1993;121:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 52.Buhrmester H, von Kries J P, Stratling W H. Biochemistry. 1995;34:4108–4117. doi: 10.1021/bi00012a029. [DOI] [PubMed] [Google Scholar]
- 53.von Kries J P, Buhrmester H, Stratling W H. Cell. 1991;64:123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]
- 54.Luderus M E E, de Graaf A, Mattia E, den Blaauwen J L, Grande M A, de Jong L, van Driel R. Cell. 1992;70:949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- 55.Luderus M E E, den Blaauwen J L, de Smit O J B, Compton D A, van Driel R. Mol Cell Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo B, Odgren P R, van Wijnen A J, Last T J, Nickerson J, Penman S, Lian J B, Stein J L, Stein G S. Proc Natl Acad Sci USA. 1995;92:10526–10530. doi: 10.1073/pnas.92.23.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 58.Zhang D E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussell M F, Gilliland D G, Lenny N, Meyers S. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 62.Banarjee, C., McCabe, L., Choi, J., Hiebert, S., Stein, J., Stein, G. & Lian, J. (1997) J. Cell. Biochem., in press. [DOI] [PubMed]
- 63.Lindenmuth, D., van Wijnen, A., Hiebert, S., Stein, J., Lian, J. & Stein, G. (1997) J. Cell. Biochem., in press. [PubMed]