Abstract
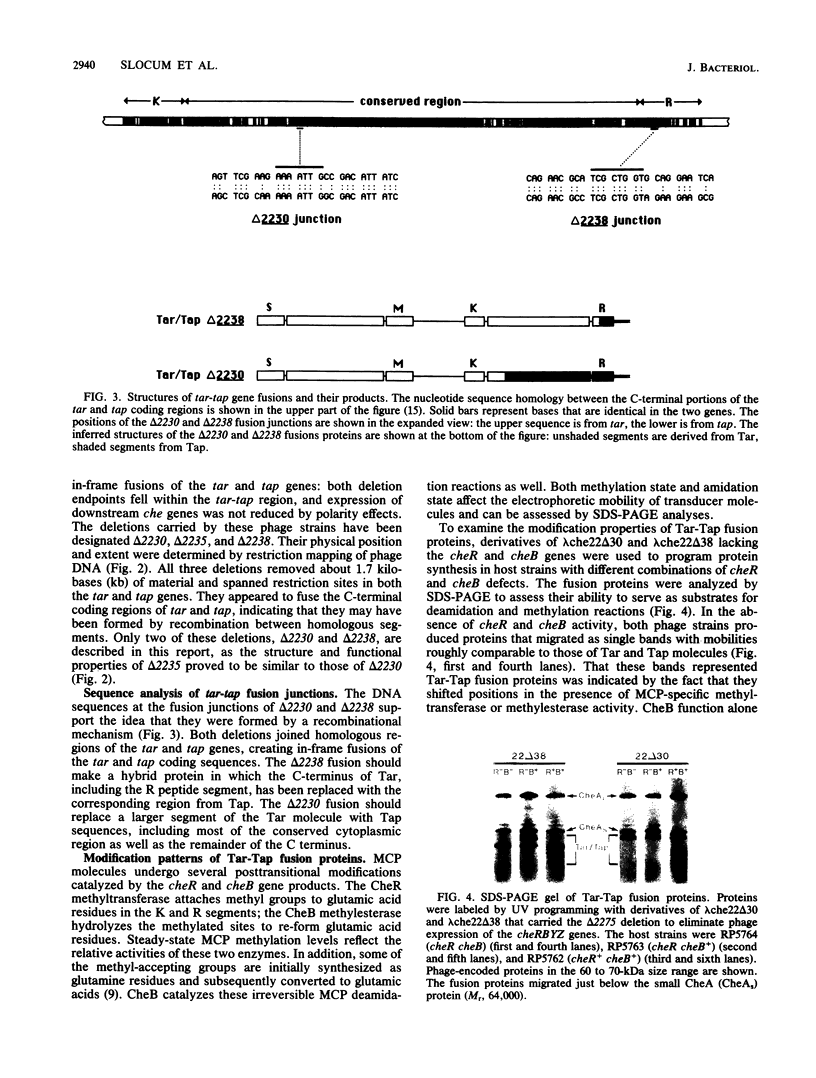
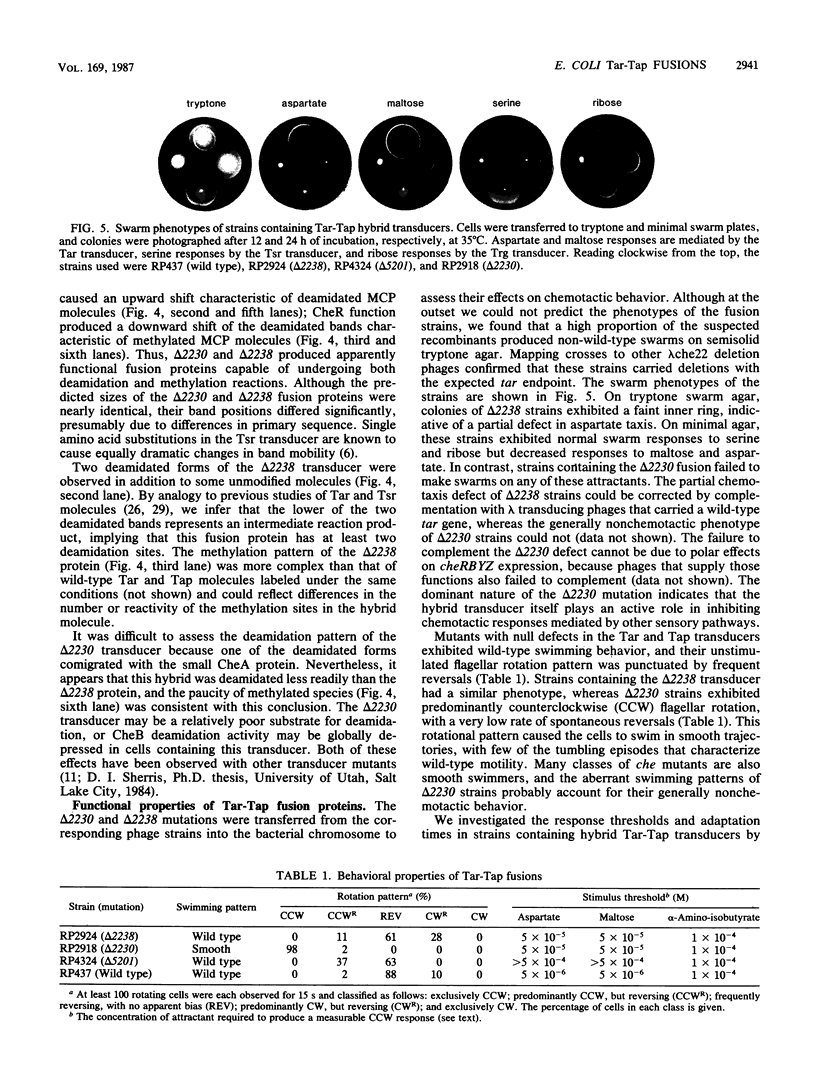
The tar and tap loci of Escherichia coli encode methyl-accepting inner membrane proteins that mediate chemotactic responses to aspartate and maltose or to dipeptides. These genes lie adjacent to each other in the same orientation on the chromosome and have extensive sequence homology throughout the C-terminal portions of their coding regions. Many spontaneous deletions in the tar-tap region appear to be generated by recombination between these regions of homology, leading to gene fusions that produce hybrid transducer molecules in which the N terminus of Tar is joined to the C terminus of Tap. The properties of two such hybrids are described in this report. Although Tar and Tap molecules have homologous domain structures, these Tar-Tap hybrids exhibited defects in stimulus detection and flagellar signaling. Both hybrid transducers retained Tar receptor specificity, but had reduced detection sensitivity. This defect was correlated with the presence of the C-terminal methyl-accepting segment of Tap, which may have more methylation sites than its Tar counterpart, leading to elevated steady-state methylation levels in the hybrid molecules. One of the hybrids, which carried a more extensive segment from Tap, appeared to generate constitutive signals that locked the flagellar motors in a counterclockwise rotational mode. Changes in the methylation state of this transducer were ineffective in cancelling this aberrant signal. These findings implicate the conserved C-terminal domain of bacterial transducers in the generation or regulation of flagellar signals.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg H. C., Tedesco P. M. Transient response to chemotactic stimuli in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3235–3239. doi: 10.1073/pnas.72.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Boyd A., Krikos A., Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981 Nov;26(3 Pt 1):333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- Callahan A. M., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J Bacteriol. 1985 Jan;161(1):96–104. doi: 10.1128/jb.161.1.96-104.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Bond M. W., Hunkapiller M. W., Dahlquist F. W. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J Biol Chem. 1982 Sep 10;257(17):10378–10386. [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Aberrant regulation of methylesterase activity in cheD chemotaxis mutants of Escherichia coli. J Bacteriol. 1985 Jan;161(1):105–112. doi: 10.1128/jb.161.1.105-112.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Sensory adaptation in bacterial chemotaxis: regulation of demethylation. J Bacteriol. 1985 Sep;163(3):983–990. doi: 10.1128/jb.163.3.983-990.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Engström P., Dahlquist F. W., Hazelbauer G. L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983 Apr 25;258(8):5050–5055. [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Blank V., Brade G., Higgins C. F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986 May 15;321(6067):253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Oosawa K., Simon M. I. Characterization of Escherichia coli chemotaxis receptor mutants with null phenotypes. J Bacteriol. 1986 Sep;167(3):992–998. doi: 10.1128/jb.167.3.992-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutation plus amplification of a transducer gene disrupts general chemotactic behavior in Escherichia coli. J Bacteriol. 1986 Dec;168(3):1378–1383. doi: 10.1128/jb.168.3.1378-1383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978 Jul;135(1):45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins C., Dahlquist F. W. The methyl-accepting chemotaxis proteins of E. coli: a repellent-stimulated, covalent modification, distinct from methylation. Cell. 1981 Aug;25(2):333–340. doi: 10.1016/0092-8674(81)90051-9. [DOI] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sherris D., Parkinson J. S. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J Bacteriol. 1985 Aug;163(2):586–594. doi: 10.1128/jb.163.2.586-594.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. A., Parkinson J. S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Koshland D. E., Jr Quantitation of the sensory response in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1975 Feb;72(2):710–713. doi: 10.1073/pnas.72.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Koshland D. E., Jr Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984 Jun 25;259(12):7719–7725. [PubMed] [Google Scholar]
- Terwilliger T. C., Wang J. Y., Koshland D. E., Jr Surface structure recognized for covalent modification of the aspartate receptor in chemotaxis. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6707–6710. doi: 10.1073/pnas.83.18.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa H., Hayashi H. Desensitization by covalent modification of the chemoreceptor of Escherichia coli. FEBS Lett. 1986 Mar 17;198(1):21–24. doi: 10.1016/0014-5793(86)81176-0. [DOI] [PubMed] [Google Scholar]





