Abstract
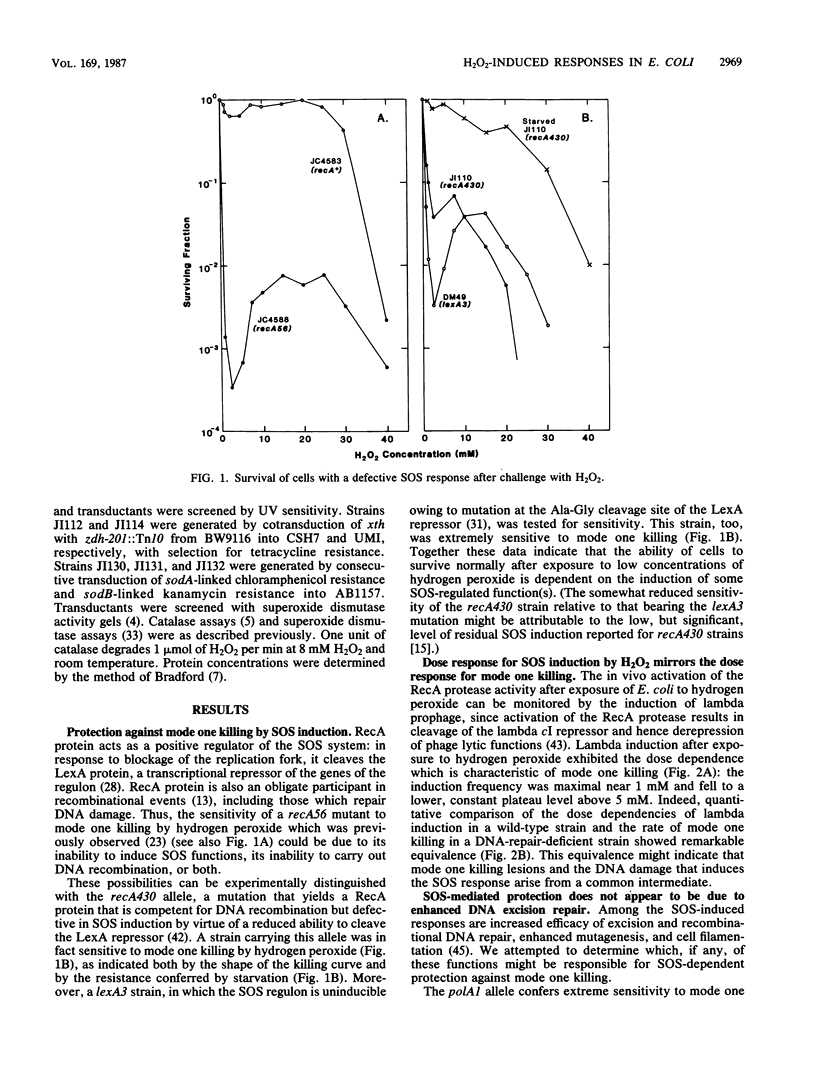
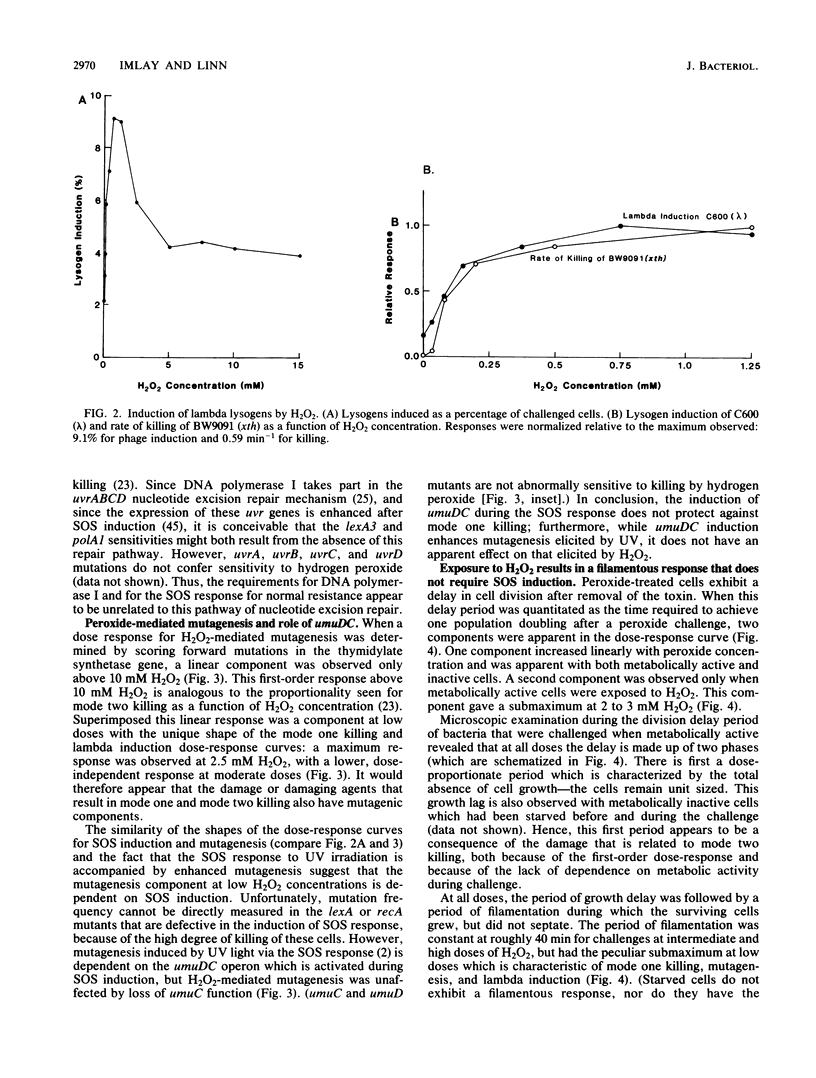
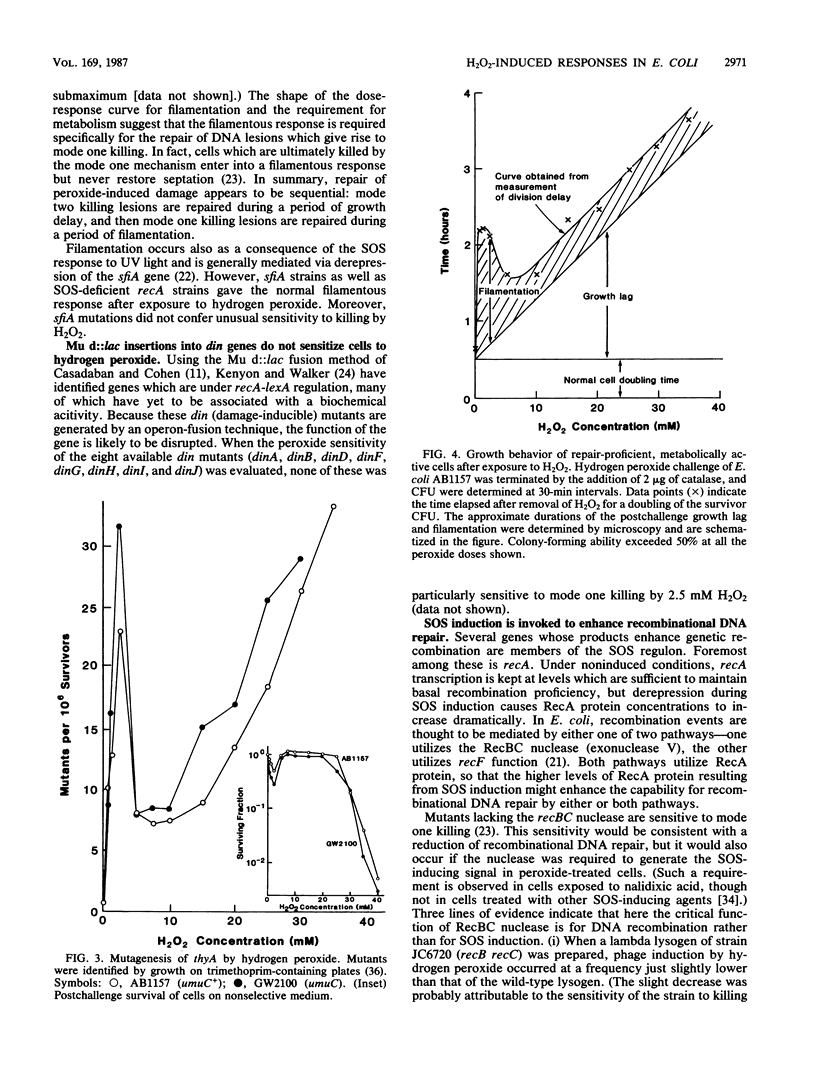
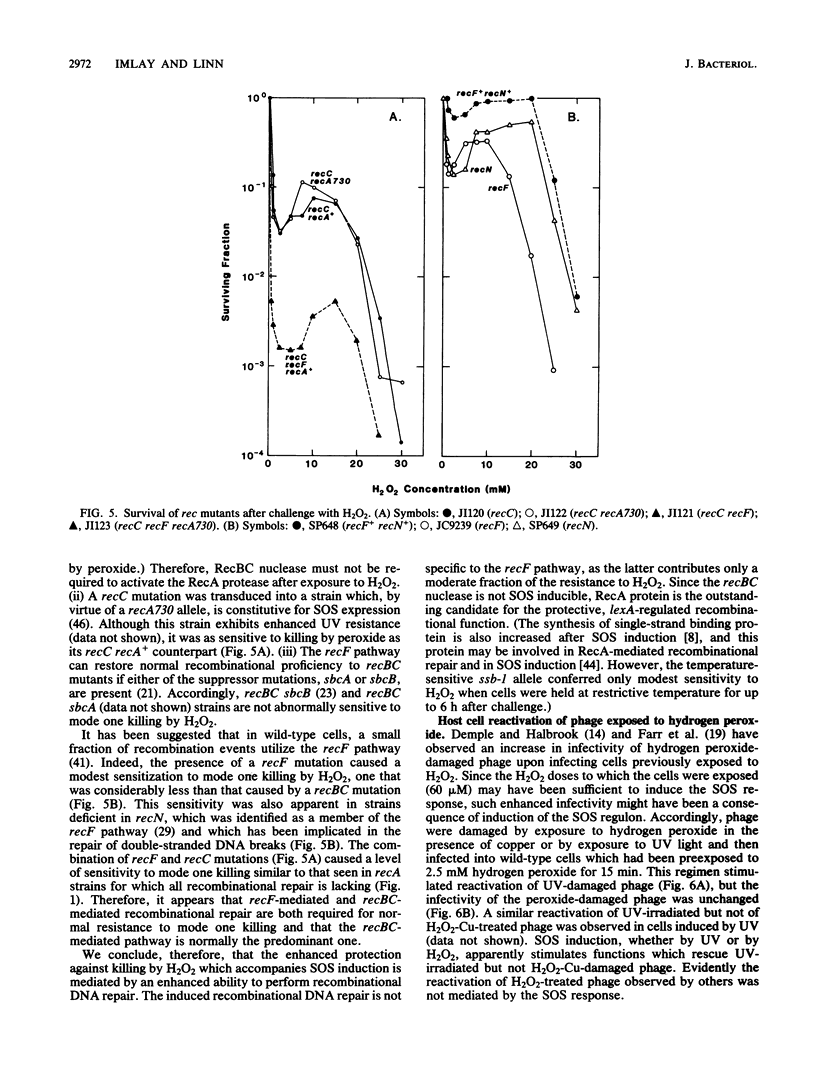
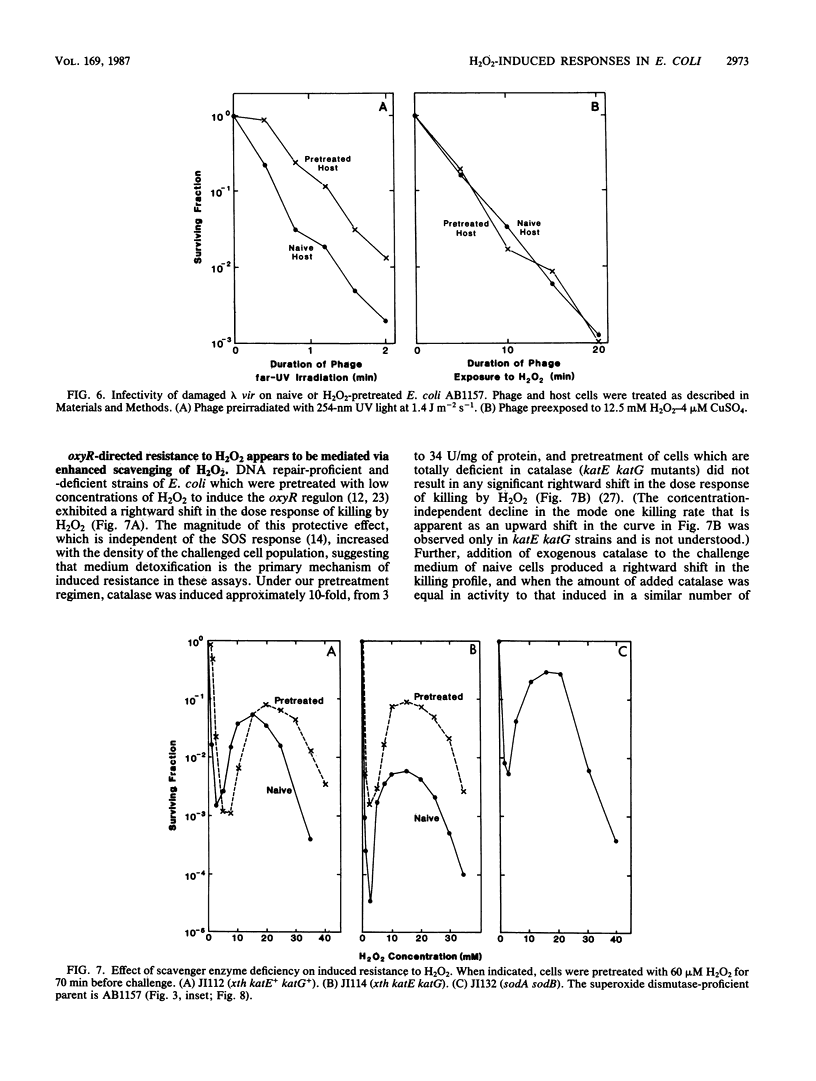
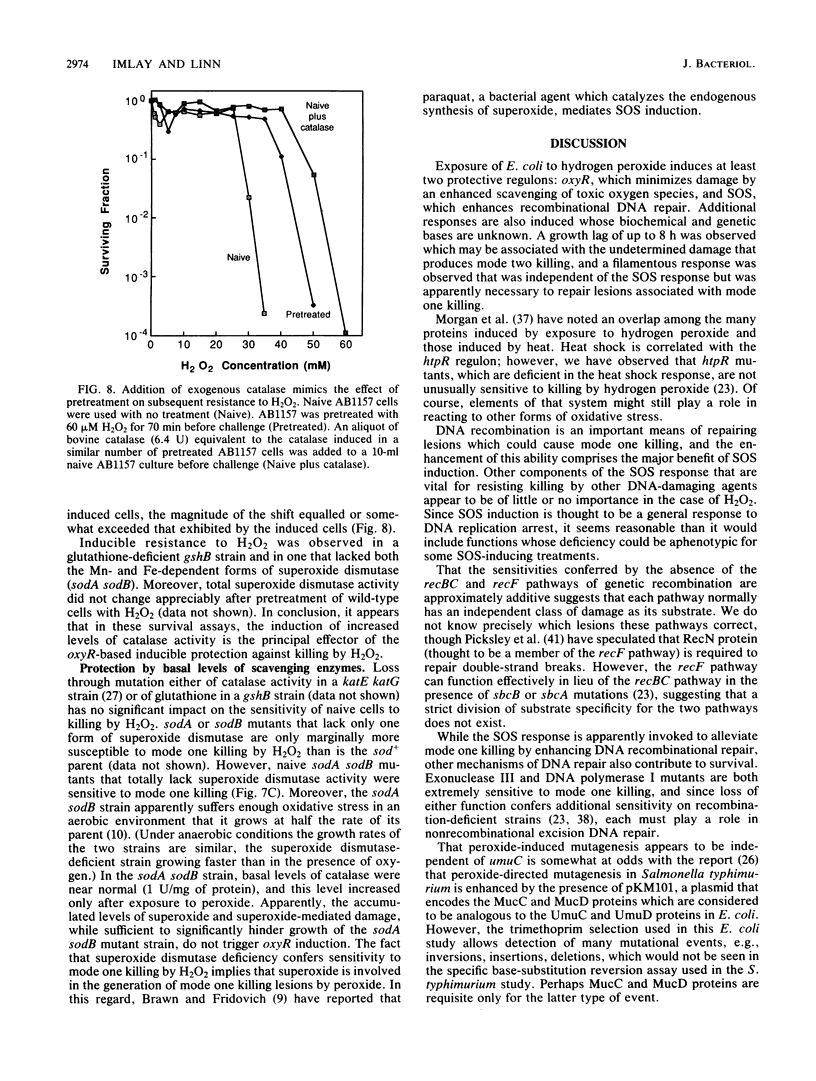
Killing of Escherichia coli by hydrogen peroxide proceeds by two modes. Mode one killing appears to be due to DNA damage, has a maximum near 1 to 3 mM H2O2, and requires active metabolism during exposure. Mode two killing is due to uncharacterized damage, occurs in the absence of metabolism, and exhibits a classical multiple-order dose-response curve up to at least 50 mM H2O2 (J. A. Imlay and S. Linn, J. Bacteriol. 166:519-527, 1986). H2O2 induces the SOS response in proportion to the degree of killing by the mode one pathway, i.e., induction is maximal after exposure to 1 to 3 mM H2O2. Mutant strains that cannot induce the SOS regulon are hypersensitive to peroxide. Analysis of the sensitivities of mutants that are deficient in individual SOS-regulated functions suggested that the SOS-mediated protection is due to the enhanced synthesis of recA protein, which is rate limiting for recombinational DNA repair. Specifically, strains wholly blocked in both SOS induction and DNA recombination were no more sensitive than mutants that are blocked in only one of these two functions, and strains carrying mutations in uvrA, -B, -C, or -D, sfiA, umuC or -D, ssb, or dinA, -B, -D, -F, -G, -H, -I, or -J were not abnormally sensitive to killing by H2O2. After exposure to H2O2, mutagenesis and filamentation also occurred with the dose response characteristic of SOS induction and mode one killing, but these responses were not dependent on the lexA-regulated umuC mutagenesis or sfiA filamentation functions, respectively. Exposure of E. coli to H2O2 also resulted in the induction of functions under control of the oxyR regulon that enhance the scavenging of active oxygen species, thereby reducing the sensitivity to H2O2. Catalase levels increased 10-fold during this induction, and katE katG mutants, which totally lack catalase, while not abnormally sensitive to killing by H2O2 in the naive state, did not exhibit the induced protective response. Protection equal to that observed during oxyR induction could be achieved by the addition of catalase to cultures of naive cells in an amount equivalent to that induced by the oxyR response. Thus, the induction of catalase is necessary and sufficient for the observed oxyR-directed resistance to killing by H2O2. Although superoxide dismutase appeared to be uninvolved in this enhanced protective response, sodA sodB mutants, which totally lack superoxide dismutase, were especially sensitive to mode one killing by H2O2 in the naive state. gshB mutants, which lack glutathione, were not abnormally sensitive to killing by H2O2.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BEERS R. F., Jr, SIZER I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952 Mar;195(1):133–140. [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagg A., Kenyon C. J., Walker G. C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour S. D., Clark A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. I. Enzymatic activity associated with recB+ and recC+ genes. Proc Natl Acad Sci U S A. 1970 Apr;65(4):955–961. doi: 10.1073/pnas.65.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp C., Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971 Nov;44(1):276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandsma J. A., Bosch D., Backendorf C., van de Putte P. A common regulatory region shared by divergently transcribed genes of the Escherichia coli SOS system. Nature. 1983 Sep 15;305(5931):243–245. doi: 10.1038/305243a0. [DOI] [PubMed] [Google Scholar]
- Brawn M. K., Fridovich I. Increased superoxide radical production evokes inducible DNA repair in Escherichia coli. J Biol Chem. 1985 Jan 25;260(2):922–925. [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A., Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986 Mar;5(3):623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Morgan R. W., Jacobson F. S., Ames B. N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985 Jul;41(3):753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- Demple B., Halbrook J. Inducible repair of oxidative DNA damage in Escherichia coli. Nature. 1983 Aug 4;304(5925):466–468. doi: 10.1038/304466a0. [DOI] [PubMed] [Google Scholar]
- Devoret R., Pierre M., Moreau P. L. Prophage phi 80 is induced in Escherichia coli K12 recA430. Mol Gen Genet. 1983;189(2):199–206. doi: 10.1007/BF00337804. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Farr S. B., D'Ari R., Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr S. B., Natvig D. O., Kogoma T. Toxicity and mutagenicity of plumbagin and the induction of a possible new DNA repair pathway in Escherichia coli. J Bacteriol. 1985 Dec;164(3):1309–1316. doi: 10.1128/jb.164.3.1309-1316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Imlay J. A., Linn S. Bimodal pattern of killing of DNA-repair-defective or anoxically grown Escherichia coli by hydrogen peroxide. J Bacteriol. 1986 May;166(2):519–527. doi: 10.1128/jb.166.2.519-527.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumura K., Sekiguchi M., Steinum A. L., Seeberg E. Stimulation of the UvrABC enzyme-catalyzed repair reactions by the UvrD protein (DNA helicase II). Nucleic Acids Res. 1985 Mar 11;13(5):1483–1492. doi: 10.1093/nar/13.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Hollstein M., Christman M. F., Schwiers E. A., Ames B. N. A new Salmonella tester strain (TA102) with A X T base pairs at the site of mutation detects oxidative mutagens. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7445–7449. doi: 10.1073/pnas.79.23.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S., Imlay J. A. Toxicity, mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Cell Sci Suppl. 1987;6:289–301. doi: 10.1242/jcs.1984.supplement_6.19. [DOI] [PubMed] [Google Scholar]
- Little J. W., Edmiston S. H., Pacelli L. Z., Mount D. W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R. G., Picksley S. M., Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Loewen P. C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984 Feb;157(2):622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham B. E., Little J. W., Mount D. W. Nucleotide sequence of the lexA gene of Escherichia coli K-12. Nucleic Acids Res. 1981 Aug 25;9(16):4149–4161. doi: 10.1093/nar/9.16.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord J. M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969 Nov 25;244(22):6049–6055. [PubMed] [Google Scholar]
- McCord J. M. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985 Jan 17;312(3):159–163. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- McPartland A., Green L., Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980 Jul;20(3):731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Milcarek C., Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972 Jul 21;68(2):303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- Morgan R. W., Christman M. F., Jacobson F. S., Storz G., Ames B. N. Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8059–8063. doi: 10.1073/pnas.83.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimyo M. Anaerobic incubation enhances the colony formation of a polA recB strain of Escherichia coli K-12. J Bacteriol. 1982 Oct;152(1):208–214. doi: 10.1128/jb.152.1.208-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount D. W., Low K. B., Edmiston S. J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet lght-induced mutations. J Bacteriol. 1972 Nov;112(2):886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang P. P., Walker G. C. Identification of the uvrD gene product of Salmonella typhimurium LT2. J Bacteriol. 1983 Mar;153(3):1172–1179. doi: 10.1128/jb.153.3.1172-1179.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley S. M., Attfield P. V., Lloyd R. G. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol Gen Genet. 1984;195(1-2):267–274. doi: 10.1007/BF00332758. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W., Craig N. L. Escherichia coli recA gene product inactivates phage lambda repressor. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4714–4718. doi: 10.1073/pnas.75.10.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Two mutations that alter the regulatory activity of E. coli recA protein. Nature. 1981 Apr 2;290(5805):422–424. doi: 10.1038/290422a0. [DOI] [PubMed] [Google Scholar]
- Vales L. D., Chase J. W., Murphy J. B. Effect of ssbA1 and lexC113 mutations on lambda prophage induction, bacteriophage growth, and cell survival. J Bacteriol. 1980 Aug;143(2):887–896. doi: 10.1128/jb.143.2.887-896.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M., McCall J. O., Volkert M. R., Wermundsen I. E. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol Gen Genet. 1982;185(1):43–50. doi: 10.1007/BF00333788. [DOI] [PubMed] [Google Scholar]



