Abstract
The hematopoietic-restricted transcription factor GATA-1 is required for both mammalian erythroid cell and megakaryocyte differentiation. To define the mechanisms governing its transcriptional regulation, we replaced upstream sequences including a DNase I hypersensitive (HS) region with a neomycin-resistance cassette by homologous recombination in mouse embryonic stem cells and generated mice either harboring this mutation (neoΔHS) or lacking the selection cassette (ΔneoΔHS). Studies of the consequences of these targeted mutations provide novel insights into GATA-1 function in erythroid cells. First, the neoΔHS mutation leads to a marked impairment in the rate or efficiency of erythroid cell maturation due to a modest (4- to 5-fold) decrease in GATA-1 expression. Hence, erythroid differentiation is dose-dependent with respect to GATA-1. Second, since expression of GATA-1 from the ΔneoΔHS allele in erythroid cells is largely restored, transcription interference imposed by the introduced cassette must account for the “knockdown” effect of the mutation. Finally, despite the potency of the upstream sequences in conferring high-level, developmentally appropriate expression of transgenes in mice, other cis-regulatory elements within the GATA-1 compensate for its absence in erythroid cells. Our work illustrates the usefulness of targeted mutations to create knockdown mutations that may uncover important quantitative contributions of gene function not revealed by conventional knockouts.
Keywords: homologous recombination, hematopoiesis, gene dosage, Cre recombinase
During the production of blood cells, pluripotent hematopoietic stem cells generate progenitors that ultimately differentiate along a single pathway (1). Precursors of erythroid cells develop from multipotential progenitors, variably committed to erythroid, megakaryocytic, and mast cell lineages. Studies aimed at understanding how a erythroid cell program of gene expression is established culminated in the identification of a consensus DNA-binding site [the GATA motif (2)] that is recognized by the hematopoietic-restricted zinc-finger transcription factor GATA-1 (3–5). This nuclear protein is abundant in several hematopoietic lineages (erythroid, mast, megakaryocytic, and eosinophil) and present at lower level in multipotential progenitors from which they arise (6). Sertoli cells of the testis are the only nonhematopoietic cells known to express GATA-1 (7).
Genetic studies in embryonic stem (ES) cells and mice established an in vivo requirement for GATA-1 in the development of both erythroid precursors and megakaryocytes (the precursors of platelets) (8–11). Disruption of the X-linked GATA-1 locus leads to embryonic lethality at the yolk sac stage due to extreme anemia (11). Primitive (i.e., yolk sac or embryonic) red cell precursors lacking GATA-1 arrest at the proerythroblast stage in vivo and appear to undergo apoptosis (11), as do adult (i.e., fetal liver or definitive) precursors generated in vitro from differentiation of GATA-1− ES cells (9). Megakaryocytes lacking GATA-1 fail to mature normally and exhibit unrestrained proliferation rather than apoptosis (10). Thus, GATA-1 function is required for cellular maturation as well as for the proper balance between proliferation, cell death, and differentiation in two related hematopoietic lineages.
Of particular interest is how GATA-1 acts to control transcription in erythroid cells. Although GATA-1 functions as a potent transcriptional activator of simple reporter constructs in nonhematopoietic cells (12, 13), GATA-1 lacking the “essential” N-terminal activation domain remains fully active in promoting terminal erythroid maturation (14). These observations and others (15, 16) suggest that the function of GATA-1 relies on the presence of an associated cofactor (14). Support for this model has been provided by the isolation of an interacting protein (FOG) (17). Thus, the activity of GATA-1 is regulated in part through critical protein–protein interactions.
In considering how GATA-1 acts in hematopoietic cells, we have also sought to define mechanisms governing the transcriptional control of the GATA-1 gene. In this regard, we recently identified a DNase I hypersensitive (HS) region upstream of the erythroid GATA-1 promoter that confers high-level, developmentally appropriate transgene expression in mice (M.A.M., Y.F., R.A.S., and S.H.O., unpublished data). To determine the in situ role of these putative regulatory sequences we replaced the HS region (and neighboring sequences) with a neomycin-resistance cassette by homologous recombination in ES cells and examined the consequences both in vitro and in vivo (10). As described here, these experiments provide novel insights into GATA-1 function and regulation in erythroid cells. Replacement of the HS region by the neomycin cassette leads to reduced levels of GATA-1 mRNA and protein—a “knockdown” mutation at the GATA-1 locus. The principal effect of this mutation is a slower rate or efficiency of erythroid cell maturation. Hence, differentiation is dose-dependent with respect to GATA-1. We also show that removal of the selection cassette by Cre-mediated site-specific recombination (18) in ES cells restores GATA-1 expression in erythroid cells, thereby providing evidence for transcriptional interference imposed by the introduced cassette (19–22). Our studies demonstrate that, despite the potency of the upstream HS region in conferring efficient and cell-restricted transgene expression in vivo, other cis-regulatory elements normally present in the GATA-1 locus compensate for its absence in erythroid precursor cells.
MATERIALS AND METHODS
neoΔHS and ΔneoΔHS ES Cells and Mice.
The generation of ES cells and mice has been described (10). The structures of the modified loci were confirmed by Southern blot analysis (see ref. 10) and by genomic PCR analysis using primer pairs surrounding the IT exon, the neomycin-resistance cassette, and the single loxP site of the ΔneoΔHS locus (data not shown). Primer pairs used were as follows: neomycin-resistance cassette, GCCCGGTTCTTTTTGTCAAGACCG and CAGAAGAACTCGTCAAGAAGGCGA; IT exon, ACTCTTGCTCTCTTTTGCAG and AATCAGGAATGCAACATCTC; and sequences flanking the loxP site, GTGTGAGAGTGGCTATGTGC and GACCCATCCATCTCCTTTCC.
Hematological Analyses.
Blood counts were obtained on a Technicon H1 or H3 analyzer. Newborn blood was diluted in acid/citrate/dextrose to obtain adequate volumes to perform automated analysis. Peripheral blood smears were stained with May-Grunwald-Giemsa.
Cell Culture and Colony Assays.
In vitro cultures of single cell suspensions of embryonic day (E) 11.5–13.5 fetal liver cells were as described (23). In vitro differentiation of ES cells was assessed by a two-step procedure (9, 24).
Detection of GATA-1 RNA and Protein.
GATA-1 RNA transcripts were assessed by semiquantitative reverse transcription–PCR (RT-PCR) using total RNA isolated from erythroid colonies or fetal liver erythroid cells. RT-PCR was performed as described (9, 24) using primer pairs specific for murine GATA-1 and a housekeeping transcript, hypoxanthine phosphoribosyltransferase. Relative transcript levels were determined by PhosphorImager (Molecular Dynamics) analysis. Nuclear extracts were prepared from fetal liver cells as described (25). Samples (2.5 μg per lane) were subjected to Western blot analysis (26) using anti-GATA-1 monoclonal N6 antibody (Santa Cruz Biotechnology). Filters were reprobed with rabbit antibody specific for transcription factor BKLF (26) as a control for protein loading.
RESULTS
Targeted Replacement of the Upstream HS Site Region by Homologous Recombination in ES Cells.
The upstream region of the GATA-1 locus contains two short noncoding 5′-exons and their associated promoters (IT for I-testes and IE for I-erythroid) and a DNase I HS site region (Fig. 1). Homologous recombination was employed to generate ES cells and mice harboring two GATA-1 alleles, designated neoΔHS and ΔneoΔHS (Fig. 1) as described (10). In the neoΔHS locus a neomycin-resistance cassette flanked by loxP recognition sites replaces ≈8 kb of upstream sequences including the HS region. To eliminate transcriptional effects of introduced sequences, Cre recombinase-mediated excision of the selection cassette (18) was performed, yielding the ΔneoΔHS allele in which only a single loxP site remained. Southern blot analyses and genomic PCR assays established the structure of the modified GATA-1 loci in ES cells and mice (data not shown; ref. 10).
Figure 1.
Structures of wild-type, neoΔHS, and ΔneoΔHS GATA-1 alleles. The wild-type locus contains alternative first exons (IT and IE) and a DNase I HS region (10). Exons are indicated by solid boxes. In the neoΔHS locus a loxP-flanked neomycin-resistance cassette replaces ≈8 kb lying between a BamHI site (B) and an artificial XhoI site (〈Xh〉) present in a λ phage genomic clone used in the construction. Cre-mediated excision of the neomycin-resistance cassette leaves a single loxP site (indicated by the solid triangle) in the ΔneoΔHS locus.
neoΔHS, But Not ΔneoΔHS, Mice Are Anemic and Exhibit Poor Viability.
Female mice heterozygous for either the neoΔHS and ΔneoΔHS alleles were mated separately with normal males. Among liveborn pups, males harboring the neoΔHS allele were greatly underrepresented (2% versus expected 25%), whereas ΔneoΔHS mice were present at near the expected frequency (Table 1). Of the few affected neoΔHS pups obtained, most were noted to be very pale at birth; ≈50% died within the first 48 hr postpartum (data not shown). Hemoglobin levels and hematocrits of 1- to 2-day-old neoΔHS males were 6.1 ± 1.1 and 29.0 ± 2.6, as compared with 11 ± 0.8 and 41.6 ± 6.6 for controls and 8.7 ± 0.6 and 31.7 ± 2.7 for heterozygous females, respectively. Remarkably, pallor and anemia evident at birth disappeared by 4–5 weeks of age. Peripheral blood smears paralleled these changes. Blood smears of newborns revealed abnormal circulating nucleated erythroid precursors and erythrocytes. Adult erythroid cell morphology was unremarkable (data not shown). Surviving male neoΔHS mice are fertile and have an apparently normal lifespan. ΔneoΔHS mice are normally viable and not anemic.
Table 1.
Genotypes of neoΔHS and ΔneoΔHS matings
| Age
|
|||||
|---|---|---|---|---|---|
| Postnatal | E13.514.5 | E9.512.5 | Adult | E14.5 | |
| neoΔHS matings | |||||
| Total (no. of litters) | 233 (33) | 156 (38) | 89 (10) | ||
| Males | |||||
| wt (%) | 87 (37) | 34 | 22 | ||
| neoΔHS (%) | 5 (2) | 34* | 18 | ||
| Females | |||||
| wt/wt (%) | 52 (22) | 44 | 24 | ||
| wt/neoΔHS | 89 (38) | 24 | 25 | ||
| ΔneoΔHS matings | |||||
| Total (no. of litters) | 89 (10) | 21 (2) | |||
| Males | |||||
| wt (%) | 22 (25) | 4 | |||
| ΔneoΔHS (%) | 18 (20) | 9 | |||
| Females | |||||
| wt/wt (%) | 24 (30) | 6 | |||
| wt/ΔneoΔHS (%) | 25 (24) | 2 | |||
Heterozygous females were mated with wild-type (wt) C57B1/6 males.
Five embryos were dead.
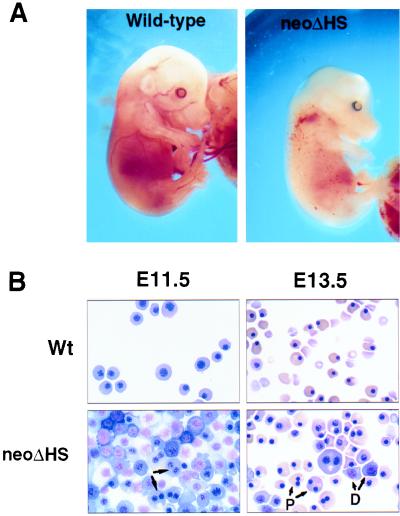
Embryos were also examined at various gestational ages (Table 1). Whereas neoΔHS and ΔneoΔHS embryos were found at the expected frequencies at the yolk sac stage, neoΔHS embryos were typically pale (Fig. 2A) or dead by E13.5–14.5 (Table 1). neoΔHS embryos were not grossly abnormal at the yolk sac stage (E9.5–12.5); ΔneoΔHS embryos appeared normal at all stages (data not shown).
Figure 2.
Anemia and abnormal erythropoiesis in neoΔHS embryos. (A) Embryos at E13.5. (B) Peripheral blood smears at yolk sac (E11.5) and fetal liver (E13.5) stages. Note binucleate primitive erythrocytes, indicated by the arrows on the left and arrows with P on the right. Note the presence of immature definitive erythroid cells (arrows with D) at E13.5.
Erythroid Maturation Is Abnormal in neoΔHS Mice.
Erythropoiesis in neoΔHS embryos was perturbed at both yolk sac and fetal liver stages of development, as shown by peripheral blood smears (Fig. 2B). Primitive erythropoiesis was dysplastic, characterized by mildly retarded cellular maturation and reduced hemoglobin accumulation and frequent binucleate erythrocytes (Fig. 2B). At the fetal liver stage (E13.5) circulating definitive erythroid cells were largely arrested at the proerythroblast stage of their development; few adult erythrocytes were evident. Numerous binucleate primitive erythrocytes were also present (Fig. 2B). The total number of fetal liver cells was reduced ≈50% relative to control littermates (data not shown).
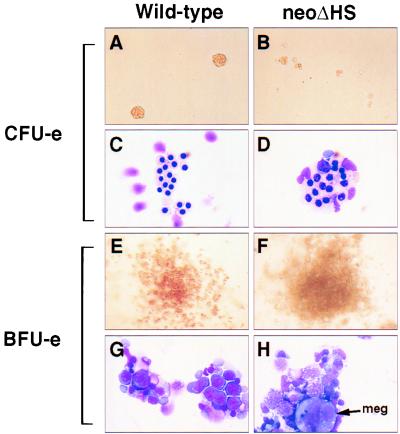
To assess hematopoietic progenitors, colony assays were performed using fetal liver cells of both neoΔHS and ΔneoΔHS embryos. As summarized in Table 2, the frequencies of early and late type erythroid colonies, known as burst-forming unit erythroid (BFU-e) and colony-forming unit erythroid (CFU-e), respectively, were not markedly different from wild-type or heterozygous controls. Although the frequency of neoΔHS erythroid colonies was essentially normal, their overall appearance, and the cells contained within them, were abnormal (Fig. 3). CFU-e were greatly reduced in size and contained proerythroblasts, as well as more mature erythroid cells, characterized by a nuclear/cytoplasmic ratio larger than that of normal late normoblasts (Fig. 3 A–D). BFU-e were abnormally large and appeared paler in color (Fig. 3 E and F). Immature erythroid cells, dysplastic and dying proerythroblasts and increased numbers of megakaryocytes were prominent (Fig. 3 G and H). Fetal liver BFU-e and CFU-e of ΔneoΔHS mice were normal in number (Table 2) and appearance (data not shown). Our findings establish that the severe anemia seen in neoΔHS embryos results from a primary defect in the maturation of erythroid precursors rather than a decreased number of erythroid progenitors.
Table 2.
Hematopoietic colony assays of neoΔHS and ΔneoΔHS fetal liver cells
| Colony type
|
|||||
|---|---|---|---|---|---|
| CFU-e | BFU-e | CFU-GM | CFU-e | BFU-e, mixed | |
| neoΔHS | |||||
| wt/wt | 4,800 ± 1,200 | 56 ± 15 | 360 ± 21 | ||
| wt/neoΔHS | 2,800 ± 450 | 63 ± 9 | 500 ± 69 | ||
| neoΔHS* | 3,300 ± 820 | 38 ± 12 | 620 ± 93 | ||
| ΔneoΔHS | |||||
| wt/wt | 3,100 ± 840 | 29 ± 7 | |||
| wt/ΔneoΔHS | 2,800 ± 1,100 | 32 ± 6 | |||
| ΔneoΔHS* | 2,500 ± 620 | 34 ± 6 | |||
Fetal liver cells were cultured as described in Materials and Methods.
Hemizygous males.
Figure 3.
Impaired erythroid colony formation of neoΔHS fetal liver cells. CFU-e and BFU-e colonies were cultured as described. Note the small size and pallor of mutant CFU-e colonies (A and B) and the increased nuclear/cytoplasmic ratio of stained erythroid cells (C and D). neoΔHS BFU-e are larger in size and less red in appearance (E and F). BFU-e contain less mature and dysplastic erythroid precursors and abundant megakaryocytes (G and H).
The neoΔHS Allele Is a Gene “Knockdown”.
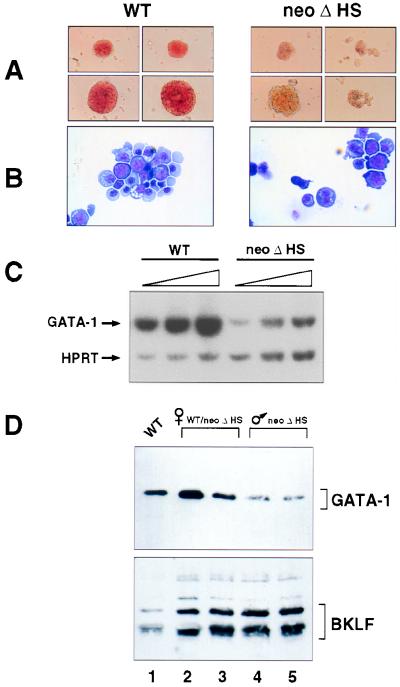
The phenotype of neoΔHS embryos is far less severe than that seen in GATA-1− embryos, which invariably die by E11 due to proerythroblast arrest and apoptosis of primitive erythroid precursors (11). We surmised, therefore, that we had created a knockdown mutation and sought to determine the level of expression. First, we assessed maturation and GATA-1 RNA transcript levels in definitive erythroid colonies obtained by in vitro differentiation of wild-type and neoΔHS ES cells. Colonies of neoΔHS origin were less robust and paler in appearance than wild-type controls and exhibited delayed erythroid cell maturation (Fig. 4 A and B). These findings recapitulate the behavior of fetal liver erythroid colonies obtained from mice (Fig. 3). GATA-1 transcripts levels in pools of definitive neoΔHS erythroid colonies were 4.7-fold reduced compared with wild type, as shown by semiquantitative PT-PCR analysis (Fig. 4C). Analysis of primitive erythroid colonies obtained by in vitro ES cell differentiation revealed a similarly modest deficiency of GATA-1 RNA (data not shown). Consistent with the presence of appreciable GATA-1 RNA, nuclear GATA-1 protein was detected in neoΔHS erythroid cells by immunofluorescence (not shown). Similar analyses of ΔneoΔHS erythroid cells failed to reveal any appreciable deficiency of RNA or protein (data not shown).
Figure 4.
Decreased GATA-1 RNA and protein levels in neoΔHS erythroid cells. (A and B) Appearance of representative definitive erythroid colonies and cells obtained by in vitro differentiation of ES cells. Colonies were harvested at day 5 after disaggregation of embryoid bodies. Note the pallor of mutant colonies and the predominance of immature erythroid cells. (C) Semiquantitative RT-PCR analysis of GATA-1 RNA transcripts. Assays were performed as described (9) with 28, 30, and 32 cycles of PCR (shown by triangles) and primer pairs specific for GATA-1 and hypoxanthine phosphoribosyltransferase (HPRT) (as an internal control). Relative band intensities were quantitated by PhosphorImager analysis. (D). Western blot analysis of fetal liver erythroid cell nuclear proteins. Wild-type female (lane 1), heterozygous neoΔHS female (lanes 2 and 3), and hemizygous neoΔHS males (lanes 4 and 5) are indicated.
To examine the level of GATA-1 protein in vivo we performed Western blot analysis of nuclear extracts of E13.5 fetal liver cells. As shown in Fig. 4D, GATA-1 protein was decreased ≈3- to 5-fold in abundance (relative to the transcription factor BKLF) in hemizygous neoΔHS males as compared with littermate controls. Thus, in vitro and in vivo neoΔHS erythroid cells express a reduced, yet readily detectable, amount of GATA-1. Similar analyses of ΔneoΔHS erythroid cells failed to reveal an appreciable deficit of GATA-1 protein (data not shown).
DISCUSSION
Our studies of the hematopoietic-specific transcription factor GATA-1 provide unique insights into the control of erythroid cell differentiation. In addition, they illustrate the utility of gene targeting of cis-regulatory elements for the generation of partial loss-of-function, or knockdown alleles.
Concentration-Dependent Erythroid Maturation.
Since its discovery through DNA-binding assays (2, 4) GATA-1 has been recognized to be an abundant nuclear protein, rather than a transcription factor present in minute quantities in the cell. Whether this aspect of GATA-1 control is critical to the erythroid differentiation program has been obscure. Is the amount of GATA-1 limiting for erythroid maturation? If so, what might this suggest about the mechanism by which GATA-1 acts? Prior data correlate increased GATA-1 levels with possible biological effects. First, GATA-1 is expressed at low levels in multipotential progenitor cells and accumulates to high levels in committed erythroid precursors, consistent with a role for increased GATA-1 protein in terminal maturation (6). Second, GATA-1 may act positively at its own promoter through high-affinity GATA-binding sites (27–29). Such autoregulation might function to “lock in” the differentiated state. Finally, forced expression of GATA-1 cDNA in retrovirally transformed chicken progenitors promotes development along three lineages (erythroid, thromboblast, and eosinophil), apparently in a concentration-dependent fashion (30).
By creating the neoΔHS allele, which is expressed at ≈20% of the wild-type level in erythroid precursors, we have been able to address the extent to which erythroid cell maturation depends on the abundance of GATA-1. Remarkably, erythroid maturation was perturbed in several respects. Though less affected than definitive (or adult) erythropoiesis, primitive (yolk sac) erythropoiesis was abnormal. Primitive erythrocytes were dysplastic, characterized principally by frequent binuclearity. Although the significance of this latter finding is uncertain, it may reflect an imbalance between cytoplasmic and nuclear events controlled by GATA-1. Moreover, hemoglobin accumulation in primitive erythroid cells appeared to be decreased, although the majority of embryos at the yolk sac stage seemed grossly normal. Definitive erythropoiesis was more markedly perturbed. Cellular maturation was nearly arrested at the proerythroblast stage, although the block to differentiation was leaky and variable. The vast majority of embryos died at the fetal liver stage due to this deficit. Some survived to birth, at which time they were noted to be anemic. Nonetheless, rare neoΔHS mice recovered and survived to adulthood. To account for this variability and the recovery of liveborn animals, we postulate that mean expression of mRNA from the GATA-1 locus within individual clones of erythroid progenitors is displaced to a lower level in cells harboring the neoΔHS allele. Accordingly, recovery from neonatal anemia reflects the in vivo selection of hematopoietic progenitors destined to express the highest levels of GATA-1. Selection must be exerted at the progenitor stage, rather than in maturing precursors; otherwise, anemia would not be corrected at steady-state. Although concentration-dependent reprogramming of hematopoietic lineages has been described in transformed chicken progenitors expressing GATA-1 (30), we have failed to observe unequivocal shifts in the proportion of various hematopoietic lineages in the face of absent (11) or reduced GATA-1. This may reflect compensatory mechanisms operative in vivo.
What might account for the critical dependence of the rate or efficiency of erythroid maturation on the level of GATA-1? Two possibilities may underlie our observations. First, abundant GATA-1 protein might be needed to saturate available GATA-binding sites in relevant target genes. If important target cis-elements contained GATA-sites of relatively low affinity, the extent of transcription necessary to complete the differentiation program might be achieved only at high protein concentrations. Alternatively, the requirement for abundant GATA-1 might reflect the relatively inefficient assembly of multiprotein complexes, which are likely to rely on individually weak protein–protein interactions for their stability. We speculate that high levels of GATA-1 are necessary to drive free GATA-1 into complexes with its cofactor, FOG (17), and perhaps with other interacting zinc-finger proteins, such as Sp1 (31), EKLF (31), and Rbtn2/Lmo2 (32).
Knockdown Mutation by cis-Element Gene Targeting.
Our experiments underscore the value of modifying cis-regulatory sequences in situ for the purpose of manipulating the output from a gene locus. While replacement of the HS region and neighboring upstream sequences with a neomycin-resistance cassette selectively ablates GATA-1 expression in megakaryocytes (10), the effect on expression in erythroid precursors is more modest (i.e., only 4- to 5-fold). By producing a knockdown mutation, we have revealed the concentration-dependent role of GATA-1 in terminal erythroid maturation. Restoration of GATA-1 expression and erythroid cell maturation upon removal of the neomycin-selection cassette via Cre recombinase-mediated excision (18) demonstrates that the major effect of the neoΔHS mutation on GATA-1 expression is due to the introduction of a transcriptionally active element into the chromatin domain, a form of transcriptional interference occasionally observed in other gene targeted loci (19–22). Of interest, replacement of the IT exon alone with the same neomycin-resistance cassette fails to perturb expression from the downstream IE promoter (cited in ref. 10). Therefore, the position of the introduced cassette relative to other elements within the GATA-1 locus determines the extent of promoter interference.
Apparently normal erythroid differentiation in ΔneoΔHS mice indicates that the HS region is not strictly required in vivo for GATA-1 gene transcription. It is likely that other cis-regulatory elements remaining in situ within the locus compensate for its absence. Thus, transcriptional control of the GATA-1 gene within erythroid precursors is complex and mediated by multiple elements. The full repertoire of these elements remains to be defined.
Combinatorial mechanisms for transcriptional control are likely to rely not only on the mere presence or absence of specific components, but also on their relative abundances at different stages of development or cellular maturation. The findings reported here provide a clear example of this additional level of regulation. Our work suggests that it is important to consider more closely the extent to which the levels of critical regulatory proteins influence cellular differentiation. As typical knockout experiments overlook such quantitative contributions, targeting of cis-regulatory elements may provide a useful strategy with which to study gene dosage effects in vivo.
Acknowledgments
R.A.S. and Y.F. contributed equally to this work. We thank Karen Rockwell, Carole Brown, Kerrianne Cunniff, and Sabra Goff for excellent technical assistance. M.A.M. received support from an National Institutes of Health Training Grant to the Division of Hematology, Brigham and Women’s Hospital, and a Howard Hughes Medical Institute Postdoctoral Fellowship. S.H.O. is an Investigator of the Howard Hughes Medical Institute. These studies were supported in part by a grant from the National Institutes of Health to S.H.O.
ABBREVIATIONS
- ES
embryonic stem
- HS
hypersensitive
- RT-PCR
reverse transcription–PCR
- E
embryonic day
- BFU-e
burst-forming unit erythroid
- CFU-e
colony-forming unit erythroid
References
- 1.Orkin S H. Curr Opin Genet Dev. 1996;6:597–602. doi: 10.1016/s0959-437x(96)80089-x. [DOI] [PubMed] [Google Scholar]
- 2.Evans T, Reitman M, Felsenfeld G. Proc Natl Acad Sci USA. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans T, Felsenfeld G. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 4.Martin D I K, Tsai S-F, Orkin S H. Nature (London) 1989;338:435–438. doi: 10.1038/338435a0. [DOI] [PubMed] [Google Scholar]
- 5.Tsai S F, Martin D I, Zon L I, D’Andrea A D, Wong G G, Orkin S H. Nature (London) 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 6.Orkin S H. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 7.Ito E, Toki T, Ishihara H, Ohtani H, Gu L, Yokoyama M, Engel J D, Yamamoto M. Nature (London) 1993;362:466–468. doi: 10.1038/362466a0. [DOI] [PubMed] [Google Scholar]
- 8.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S-F, D’Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 9.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 10.Shivdasani, R. A., Fujiwara, Y., McDevitt, M. A. & Orkin, S. H. (1997) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 11.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Proc Natl Acad Sci USA. 1996;93:12355–122358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin D I K, Orkin S H. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 13.Evans T, Felsenfeld G. Mol Cell Biol. 1991;11:843–853. doi: 10.1128/mcb.11.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss M J, Yu C, Orkin S H. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blobel G A, Simon M C, Orkin S H. Mol Cell Biol. 1995;15:626–633. doi: 10.1128/mcb.15.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visvader J E, Crossley M, Hill J, Orkin S H, Adams J M. Mol Cell Biol. 1995;15:634–641. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang, A. C., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell, in press. [DOI] [PubMed]
- 18.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 19.Kim C G, Epner E M, Forrester W C, Groudine M. Genes Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 20.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 21.Hug B A, Wessleschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson E N, Arnold H H, Rigby P W J, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 23.Perkins A C, Sharpe A H, Orkin S H. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 24.Keller G, Kennedy M, Papayannopoulou T, Wiles M V. Mol Cell Biol. 1993;13:472–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews N C, Faller D V. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin S H. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai S-F, Strauss E, Orkin S H. Genes Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- 28.Hannon R, Evans T, Felsenfeld G, Gould H. Proc Natl Acad Sci USA. 1991;88:3004–3008. doi: 10.1073/pnas.88.8.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trainor C D, Omichinski J G, Vandergon T L, Gronenborn A M, Clore G M, Felsenfeld G. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulessa H, Frampton J, Graf T. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 31.Merika M, Orkin S H. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]