Abstract
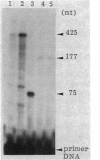
The effect of high-level expression of the regulatory gene xylS of the Pseudomonas putida TOL plasmid on the activation of the xylDLEGF operon was investigated in Escherichia coli. The xylS gene was placed downstream from the tac promoter, and the resultant fusion was cloned in cis to the xylDLEGF operon. The expression of the operon was monitored by the level of catechol 2,3-dioxygenase, whose structural gene xylE was placed directly after the operator-promoter region of xylDLEGF. xylS transcription was also determined by reverse transcriptase mapping of mRNA. Overproduction of the xylS gene product elicited constitutive high expression of the xylDLEGF operon even in the absence of the inducer for the operon. The results were consistent with a cascade model for the positive control of the xylDLEGF operon by the xylR and xylS genes (S. Inouye, A. Nakazawa, and T. Nakazawa, Proc. Natl. Acad. Sci. USA, in press): m-xylene, a substrate of the degradative pathway, binds to the xylR gene product; the m-xylene-xylR product complex activates the xylS gene; and the xylS product thus synthesized de novo activates the xylDLEGF operon.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J Bacteriol. 1982 May;150(2):722–729. doi: 10.1128/jb.150.2.722-729.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M., Clements J., Merrick M., Dixon R. Positive control and autogenous regulation of the nifLA promoter in Klebsiella pneumoniae. Nature. 1983 Jan 27;301(5898):302–307. doi: 10.1038/301302a0. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C. D., Wanner B., Inouye H. Analysis of regulation of phoB expression using a phoB-cat fusion. J Bacteriol. 1983 Nov;156(2):710–717. doi: 10.1128/jb.156.2.710-717.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the regulatory gene xylR for xyl operons on the TOL plasmid. J Bacteriol. 1985 Sep;163(3):863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J Bacteriol. 1983 Sep;155(3):1192–1199. doi: 10.1128/jb.155.3.1192-1199.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putida TOL plasmid and identification of the protein product. Gene. 1986;44(2-3):235–242. doi: 10.1016/0378-1119(86)90187-3. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., Jeenes D. J., Broda P. Characterization by molecular cloning of insertion mutants in TOL catabolic functions. Plasmid. 1983 Mar;9(2):112–125. doi: 10.1016/0147-619x(83)90014-8. [DOI] [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Spooner R. A., Lindsay K., Franklin F. C. Genetic, functional and sequence analysis of the xylR and xylS regulatory genes of the TOL plasmid pWW0. J Gen Microbiol. 1986 May;132(5):1347–1358. doi: 10.1099/00221287-132-5-1347. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]