Abstract
We have developed a new research tool for the study of S-adenosylmethionine (AdoMet) metabolism by cloning the coliphage T3 AdoMet hydrolase (AdoMetase; EC 3.3.1.2) gene into the M13mp8 expression vector. The recombinant bacteriophage clones expressed an AdoMetase activity in Escherichia coli like that found in T3-infected cells. High levels of AdoMetase expression impaired AdoMet-mediated activities such as dam and dcm methylase-directed DNA modifications and the synthesis of spermidine from putrescine. Expression vectors containing the cloned AdoMetase gene thus provide an alternate approach to the use of chemical inhibitors or mutants defective in AdoMet biosynthesis to probe the effect of AdoMet limitation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Basu S., Sarkar P., Maitra U. Location, function, and nucleotide sequence of a promoter for bacteriophage T3 RNA polymerase. Proc Natl Acad Sci U S A. 1981 Jan;78(1):147–151. doi: 10.1073/pnas.78.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix J. H. Molecular aspects of the in vivo and in vitro effects of ethionine, an analog of methionine. Microbiol Rev. 1982 Sep;46(3):281–295. doi: 10.1128/mr.46.3.281-295.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraj J. A., Marinus M. G. Phenotypic reversal in dam mutants of Escherichia coli K-12 by a recombinant plasmid containing the dam+ gene. J Bacteriol. 1983 Jan;153(1):562–565. doi: 10.1128/jb.153.1.562-565.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. N., Dembinski D. R., McAllister W. T. Derivation of a restriction map of bacteriophage T3 DNA and comparison with the map of bacteriophage T7 DNA. J Virol. 1980 Jul;35(1):176–183. doi: 10.1128/jvi.35.1.176-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P. K., Studier F. W., Hamilton D. L., Yuan R. Inhibition of the type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J Mol Biol. 1985 Apr 20;182(4):567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- Beier H., Hausmann R. Genetic map of bacteriophage T3. J Virol. 1973 Aug;12(2):417–419. doi: 10.1128/jvi.12.2.417-419.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt R. T. S-Adenosyl-L-methionine-dependent macromolecule methyltransferases: potential targets for the design of chemotherapeutic agents. J Med Chem. 1980 Apr;23(4):347–357. doi: 10.1021/jm00178a001. [DOI] [PubMed] [Google Scholar]
- Brown J. E., Bailey J. N., McAllister W. T. Sequence of a region near the left end of bacteriophage T3 DNA that contains three promoters for the E. coli RNA polymerase. Nucleic Acids Res. 1986 Jun 11;14(11):4696–4696. doi: 10.1093/nar/14.11.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt J., Weisemann J., Hamilton D. L., Yuan R. Complexes formed between the restriction endonuclease EcoK and heteroduplex DNA. J Mol Biol. 1981 Dec 5;153(2):425–440. doi: 10.1016/0022-2836(81)90287-4. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr, Reed R., Taylor F. R., Jackson M. B. Properties and biosynthesis of cyclopropane fatty acids in Escherichia coli. J Bacteriol. 1979 Apr;138(1):118–121. doi: 10.1128/jb.138.1.118-121.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation--a regulatory signal in eukaryotic gene expression. J Gen Virol. 1981 Nov;57(Pt 1):1–20. doi: 10.1099/0022-1317-57-1-1. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Flores H. E., Galston A. W. Analysis of polyamines in higher plants by high performance liquid chromatography. Plant Physiol. 1982 Mar;69(3):701–706. doi: 10.1104/pp.69.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H., Sugimoto K. On the terminally redundant sequences of bacteriophage T3 DNA. Virology. 1983 Jan 30;124(2):251–258. doi: 10.1016/0042-6822(83)90342-2. [DOI] [PubMed] [Google Scholar]
- GOLD M., HAUSMANN R., MAITRA U., HURWITZ J. THE ENZYMATIC METHYLATION OF RNA AND DNA. 8. EFFECTS OF BACTERIOPHAGE INFECTION ON THE ACTIVITY OF THE METHYLATING ENZYMES. Proc Natl Acad Sci U S A. 1964 Aug;52:292–297. doi: 10.1073/pnas.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Hafner E. W., Tabor C. W., Tabor H. Isolation of a metK mutant with a temperature-sensitive S-adenosylmethionine synthetase. J Bacteriol. 1977 Dec;132(3):832–840. doi: 10.1128/jb.132.3.832-840.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Schroeder C. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev. 1981 Mar;45(1):9–51. doi: 10.1128/mr.45.1.9-51.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lombardini J. B., Coulter A. W., Talalay P. Analogues of methionine as substrates and inhibitors of the methionine adenosyltransferase reaction. Deductions concerning the conformation of methionine. Mol Pharmacol. 1970 Sep;6(5):481–499. [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Pleiotropic effects of a DNA adenine methylation mutation (dam-3) in Escherichia coli K12. Mutat Res. 1975 Apr;28(1):15–26. doi: 10.1016/0027-5107(75)90309-7. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Poteete A., Arraj J. A. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984 Apr;28(1):123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- McGraw N. J., Bailey J. N., Cleaves G. R., Dembinski D. R., Gocke C. R., Joliffe L. K., MacWright R. S., McAllister W. T. Sequence and analysis of the gene for bacteriophage T3 RNA polymerase. Nucleic Acids Res. 1985 Sep 25;13(18):6753–6766. doi: 10.1093/nar/13.18.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mulligan J. T., Margolin W., Krueger J. H., Walker G. C. Mutations affecting regulation of methionine biosynthetic genes isolated by use of met-lac fusions. J Bacteriol. 1982 Aug;151(2):609–619. doi: 10.1128/jb.151.2.609-619.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E., McGill S. M. Inhibition of diamine oxidase by 1,1-[(methylethanediylidene)-dinitrilo]-bis-(3-aminoguanidine) and 1,1'-[(methylethanediylidene)-dinitrilo]-diguanidine. Biochem Pharmacol. 1978;27(12):1625–1629. doi: 10.1016/0006-2952(78)90170-3. [DOI] [PubMed] [Google Scholar]
- Peterson K. R., Wertman K. F., Mount D. W., Marinus M. G. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201(1):14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- Pine M. J. Comparative physiological effects of incorporated amino acid analogs in Escherichia coli. Antimicrob Agents Chemother. 1978 Apr;13(4):676–685. doi: 10.1128/aac.13.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Reuter M., Krüger D. H., Scholz D., Rosenthal H. A. Schutz zellfremder DNA vor wirtskontrollierter Restriktion in Bakterienzellen. II. Schutz des Plasmids pSF2124 durch die ocr+ Genfunktion der Bakteriophagen T3 und T7. Z Allg Mikrobiol. 1980;20(5):345–354. doi: 10.1002/jobm.3630200506. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes B. C., McClure W. R., Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985 Nov;43(1):117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Santi D. V., Garrett C. E., Barr P. J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983 May;33(1):9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Spoerel N., Herrlich P., Bickle T. A. A novel bacteriophage defence mechanism: the anti-restriction protein. Nature. 1979 Mar 1;278(5699):30–34. doi: 10.1038/278030a0. [DOI] [PubMed] [Google Scholar]
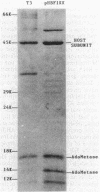
- Spoerel N., Herrlich P. Colivirus-T3-coded S-adenosylmethionine hydrolase. Eur J Biochem. 1979 Apr 2;95(2):227–233. doi: 10.1111/j.1432-1033.1979.tb12957.x. [DOI] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
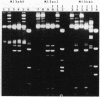
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Movva N. R. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol. 1976 Jul;19(1):136–145. doi: 10.1128/jvi.19.1.136-145.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Avraham-Haetzni K., Reifman A., Shlomai J., Kaplan F., Oppenheim A., Razin A. DNA methylation pattern is determined by the intracellular level of the methylase. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3278–3282. doi: 10.1073/pnas.81.11.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- Tabor H. Polyamine biosynthesis in Escherichia coli: construction of polyamine-deficient mutants. Med Biol. 1981 Dec;59(5-6):389–393. [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J Mol Biol. 1982 Dec 15;162(3):679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- Williams-Ashman H. G., Schenone A. Methyl glyoxal bis(guanylhydrazone) as a potent inhibitor of mammalian and yeast S-adenosylmethionine decarboxylases. Biochem Biophys Res Commun. 1972 Jan 14;46(1):288–295. doi: 10.1016/0006-291x(72)90661-4. [DOI] [PubMed] [Google Scholar]