Abstract
Natural selection can influence even the lowest level of biological organization, the atomic composition of biological macromolecules. In analysing genome-scale gene expression data, we find that ancestral yeast strains preferentially express proteins with low carbon content during carbon limitation, relative to strains selected in the laboratory under carbon limitation. The likely reason is that the artificially selected strains acquire adaptations that refine their response to the limitation or partly circumvent the limiting condition. This finding extends previous work which shows that natural selection can act on the atomic costs of proteins. We also show that genes with high carbon and nitrogen content are less likely to have duplicates, indicating that atomic composition also plays a role in evolution by gene duplication. Taken together, our results contribute to the emerging view that protein atomic composition influences genome and transcriptome evolution.
Keywords: elemental composition, proteome, evolution
1. Introduction
Natural selection affects the survival and reproduction of whole organisms, which are complex assemblages of millions of molecules. Yet its effects trickle down to the lowest levels of biological organization. Consider the amino acid components of proteins. Their biosynthesis costs energy, which is typically measured in energy equivalents of activated phosphate bonds or ∼P. The 20 proteinaceous amino acids vary in their energetic cost, in a manner that depends on biosynthetic pathways used by different organisms. Differences in the cost of amino acids have evolutionary implications (Richmond 1970). For example, in Escherichia coli, energetic costs of different amino acids vary up to sixfold, and highly expressed proteins are depleted for especially costly amino acids (Akashi & Gojobori 2002). This becomes explicable if one considers that highly expressed proteins can consume a substantial fraction of a cell's energy budget, as has been demonstrated for Saccharomyces cerevisiae (Wagner 2005). It also means that the energy savings afforded by modifying the amino acid composition of proteins are substantial enough to be visible to natural selection. Amino acid costs therefore constrain the evolution of protein composition, in addition to compositional constraints imposed by protein function and organism lifestyle (see Pascal et al. (2006) for a recent analysis and review).
The lowest level of biological organization is that of atoms in biological macromolecules. Selection's signature is even visible on this level (e.g. Mazel & Marlière 1989; Rocha et al. 2000; Baudouin-Cornu et al. 2001; Elser et al. 2006). Specifically, individual amino acids and whole proteins can vary greatly in their content of carbon, nitrogen and sulphur atoms, and this variation in elemental composition can be influenced by natural selection. The evidence ranges from anecdotal observations on individual proteins to proteome-wide patterns. For instance, Pardee (1966) observed that a sulphate-binding protein from Salmonella typhimurium is largely depleted in sulphur atoms. In whole proteomes, proteins needed to assimilate carbon are depleted in carbon atoms, and proteins needed to assimilate sulphur are depleted in sulphur atoms (Baudouin-Cornu et al. 2001). This is probably an adaptation to maintain the activity of nutrient assimilation pathways in times of nutrient scarcity (Baudouin-Cornu et al. 2001). It shows that natural selection can shape the elemental composition of specific protein classes over long evolutionary time-scales.
Gene expression data support the notion that the elemental composition of proteins has adaptive significance. Specifically, microbes can respond to sudden nutrient limitation with biases in the elemental composition of their expressed proteins. For instance, during sulphur starvation, marine bacteria produce sulphur-depleted proteins (Cuhel et al. 1981). Yeast can conserve sulphur by expressing proteins with few sulphur-containing amino acids during times of sulphur scarcity (Fauchon et al. 2002; Boer et al. 2003). The differential expression of gene duplicates (paralogues) with different sulphur content may be partly responsible for such ‘sulphur sparing’ (Mazel & Marlière 1989; Fauchon et al. 2002).
Laboratory evolution experiments have shown that prolonged glucose limitation can lead to evolutionary adaptations in gene expression after merely a few hundred generations (e.g. Ferea et al. 1999; Jansen et al. 2005). However, we do not know whether the genes whose expression changes through natural selection under carbon limitation encode proteins with biased elemental composition. With respect to protein carbon content, natural or artificial (laboratory) selection might affect gene expression in a number of possible ways. For example, it might lead to changes in transcription that reduce the carbon cost of protein expression. In other words, selection under carbon limitation could promote ‘carbon sparing’ in proteins. Artificially selected strains would then be expected to upregulate genes encoding carbon-poor proteins and to downregulate genes encoding carbon-rich proteins, relative to unselected ancestral strains. Another possibility is that selection under carbon limitation leads to exactly the opposite: a reduction in carbon sparing, relative to unselected (ancestral) strains. This could occur for several possible reasons. One is that artificially selected strains might acquire other adaptations that either refine the response to nutrient limitation or provide some relief from limitation. For example, selected strains may acquire an increased affinity for the limiting nutrient (Dykhuizen & Hartl 1981; Helling et al. 1987; Jansen et al. 2005) and may be able to access the nutrient more efficiently.
Here, we distinguish between these hypotheses. We test for biases in the carbon cost of yeast proteins whose genes were differentially expressed during low carbon availability. Specifically, we compare gene expression between strains that were artificially selected under carbon limitation and their unselected ancestral strains. We find that genes with high expression in ancestral strains do have protein products that are significantly carbon depleted. We also show that yeast genes with duplicates tend to have protein products with low carbon and nitrogen content, which demonstrates that protein elemental composition also plays a role in evolution by gene duplication.
2. Material and methods
(a) Data
We obtained translated amino acid sequences and functional annotations for predicted nuclear genes of S. cerevisiae (excluding dubious protein-coding sequences (CDSs) and pseudogenes) from the Saccharomyces Genome Database FTP site (ftp://genome-ftp.stanford.edu/pub/yeast/).
We obtained data from two studies that compared genome-scale transcript abundances of yeast growing under conditions of low glucose availability before and after artificial selection under these conditions. In the first of these studies, Ferea et al. (1999) grew three populations of S. cerevisiae for 250–500 generations in aerobic, glucose-limited conditions and used microarrays to identify CDSs whose transcript abundances were different in these artificially selected strains, relative to their ancestors. Prior to the expression analyses, all three selected populations and the ancestral strain were grown for 10–15 generations under conditions identical to those used for selection (aerobic, low glucose; Ferea et al. 1999). In our analysis, we used publicly available information on the fold change in gene expression in the selected strains and focused on genes whose expression changed by a factor of two or more, in at least two selected strains (http://genome-www.stanford.edu/evolution/; file ‘Evolall.txt’).
Our second dataset is derived from a study by Jansen et al. (2005), where S. cerevisiae were evolved for 200 generations under aerobic, glucose-limited conditions. The authors tested whether transcript levels were significantly different between the selected strain and the ancestral strain (http://www.bt.tudelft.nl/glucose-selection; files ‘up-regulated.txt’ and ‘down-regulated.txt’).
We also used data from an analysis of gene expression during physiological responses to different limiting nutrients (as opposed to comparing expression before and after selection under glucose limitation). Boer et al. (2003) performed microarray experiments to compare the abundances of transcripts from S. cerevisiae grown under conditions of limitation by carbon, nitrogen, phosphorus or sulphur. In our analysis, we focused on transcripts whose abundances were significantly different between yeast cells growing under carbon limitation, relative to yeast growing on excess carbon (but limited by N, P or S) (http://www.nutrient-limited.bt.tudelft.nl; files: ‘upC-lim.txt’ and ‘downC-lim.txt’).
The above three studies identify genes that are differentially expressed using comparisons of transcript abundances. They likely provide a good indication of which genes are differentially expressed under the experimental conditions. However, they do not consider possible determinants of protein translation rates other than transcription. This is a necessary limitation of our analyses.
(b) Duplicate genes
We identified duplicate genes in yeast using a previously published tool (Conant & Wagner 2002). This tool uses DNA and amino acid sequences of coding regions to identify related genes on a genome-wide scale in a three-step process. First, it identifies genes with sequence homology across the genome, using BLASTP (E<0.01; Altschul et al. 1997). Next, it aligns the resulting subset of genes with a global dynamic programming alignment algorithm (Thompson et al. 1994) and excludes pairs of genes that have fewer than 40 aligned amino acids or less than 50% amino acid identity.
We obtained codon adaptation index (CAI; Sharp & Li 1987) data for 5766 yeast protein-CDSs from the Saccharomyces Genome Database ftp (ftp://genome-ftp.stanford.edu/pub/yeast/).
We obtained functional categories for 5843 yeast CDSs from the MIPS Functional Catalogue (Ruepp et al. 2004; ftp://ftpmips.gsf.de/yeast/catalogues/funcat).
(c) Elemental and energetic costs
For each yeast protein, we calculated the mean (i) carbon content, (ii) nitrogen content, and (iii) energetic cost (in units of activated phosphate bonds, ‘∼P’, and reducing equivalents, ‘H’), per amino acid. We used estimates of amino acid biosynthetic costs for respiratory and fermentative conditions from Wagner (2005), except for lysine, where we take into account that yeast uses α-ketoglutarate instead of oxaloacetate as the lysine precursor. The resulting lysine biosynthesis costs are 16∼P and 1∼P for respiratory metabolism and fermentative metabolism, respectively. We test whether groups of proteins have significantly different elemental or energetic costs using Mann–Whitney U-tests. For each test, we report the p-value and the number of proteins in the two groups that are being compared (separated by a comma). In these analyses, the use of protein costs per amino acid excludes protein length as a confounding factor. This is desirable since protein length may be more constrained by functional requirements than protein amino acid and elemental composition.
3. Results
(a) No bias in protein carbon and energetic costs during physiological response
In a first analysis, we asked whether the protein products of genes whose expression is upregulated under carbon limitation (relative to their expression under N, P and S limitation; Boer et al. 2003) are depleted in carbon, as one might expect if a cell's total carbon budget is constrained. However, this is not the case. Protein products of 157 genes with higher expression under glucose limitation did not have carbon content (per amino acid) significantly different from the rest of the proteome (p=0.248, Mann–Whitney U-test, n=157, 5698). An analogous question can be asked for the energetic cost of protein expression (in units of activated phosphate bonds, ∼P), because energy production becomes limited with carbon limitation. Also, amino acids with high carbon content, such as phenylalanine, tyrosine and tryptophan, tend to have complex biosyntheses that consume more energy. In fact, carbon content (per amino acid) and mean energetic cost (per amino acid) are highly correlated among yeast proteins (respiratory: n=5855, rs=0.779, p<0.001, and fermentative: n=5855, rs=0.560, p<0.001; figure 1). The protein products of upregulated genes (identified by Boer et al. 2003) do not have significantly lower respiratory energy cost than the rest of the proteome, and in fact have a weak tendency to have greater respiratory energy costs (p=0.038, Mann–Whitney U-test, n=157, 5698). The fermentative energy costs of these proteins are not different to the rest of the proteome (p=0.578, Mann–Whitney U-test, n=157, 5698). Conversely, the protein products of genes downregulated under glucose limitation did not have significantly higher carbon content (p=0.546, Mann–Whitney U-test, n=60, 5795) or fermentative energy cost (p=0.269, Mann–Whitney U-test, n=60, 5795) than the rest of the proteome, although they did have a slightly higher respiratory energy cost (p=0.016, Mann–Whitney U-test, n=60, 5795).
Figure 1.
Relationship between the carbon content (atoms per amino acid) and energetic cost (∼P per amino acid) of yeast proteins during (a) respiratory and (b) fermentative growth.
(b) Biased protein carbon and energetic cost in short-term evolutionary adaptation
We next asked whether genes expressed at different levels in strains artificially selected (SE) under carbon limitation, relative to ancestral (AN) strains that only showed physiological responses, had protein products with significantly different carbon content from the rest of the proteome. We wanted to test whether strains that were selected under glucose limitation had evolved transcriptional responses that led to carbon sparing, or alternatively, if there was evidence for the relaxation of carbon sparing in selected strains. We used data from two laboratory selection studies of yeast, Ferea et al. (1999; abbreviated F99) and Jansen et al. (2005; abbreviated J05).
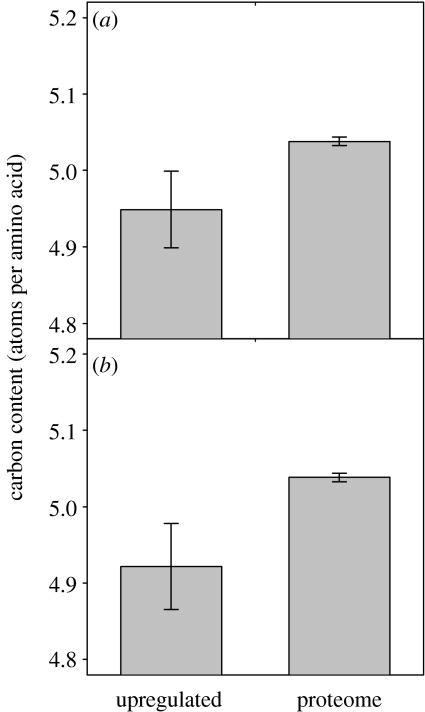
Proteins whose genes were expressed at greater levels in ancestral (AN) yeast strains relative to artificially selected (SE) strains (i.e. lower expression in selected strains relative to ancestral strains) had significantly lower carbon content than the rest of the proteome (J05: p<0.001, Mann–Whitney U-test, n=63, 5792; F99: p=0.001, Mann–Whitney U-test, n=69, 5786; figure 2a,b). We next asked whether these proteins also have lower energetic costs than the rest of the proteome. This was not the case for either fermentative energy cost (J05: p=0.146, Mann–Whitney U-test, n=63, 5792; F99: p=0.339, Mann–Whitney U-test, n=69, 5786) or respiratory energy cost (J05: p=0.606, Mann–Whitney U-test, n=63, 5792; F99: p=0.702, Mann–Whitney U-test, n=69, 5786). These analyses suggest that artificial selection did not promote carbon sparing. To the contrary, the data suggest reduced carbon sparing in selected strains, relative to ancestral strains. There was no evidence for energy sparing in either ancestral or selected strains.
Figure 2.
Mean (±2 s.e.) carbon content (atoms per amino acid) of yeast proteins whose genes had greater expression in ancestral strains responding to carbon limitation than in strains artificially selected under carbon limitation, and mean (±2 s.e.) carbon content (atoms per amino acid) of the rest of the proteins in the proteome. Data are presented for two studies, (a) Ferea et al. (1999) and (b) Jansen et al. (2005).
We wanted to know if the low carbon content of proteins whose genes had higher expression in ancestral (AN) versus selected (SE) strains was entirely attributable to a common set of proteins with gene expression AN>SE in both artificial selection studies (Ferea et al. 1999 and Jansen et al. 2005). This was not the case. There were nine genes whose expression was greater in AN than in SE strains in both Ferea et al. (1999) and Jansen et al. (2005) (out of 63 and 69 genes, respectively). Their gene products include several proteins with obvious roles in carbon metabolism, such as enolase, pyruvate decarboxylase, pyruvate kinase and glyceraldehyde-3-phosphate dehydrogenase (Saccharomyces Genome Database). When we excluded these nine proteins from our analyses, we still found that proteins encoded by genes with greater expression in ancestral (AN) than in selected (SE) strains have lower carbon content than the rest of the proteome in each study (J05: p=0.001, Mann–Whitney U-test, n=54, 5792; F99: p=0.012, Mann–Whitney U-test, n=60, 5786). Therefore, two distinct sets of proteins whose genes had greater expression in AN strains than SE strains in separate studies (Ferea et al. 1999; Jansen et al. 2005) had carbon-poor proteins.
Next, we wanted to determine whether the set of genes that had higher expression in ancestral (AN) than in artificially selected (SE) strains (in selection studies F99 and J05) overlaps with the genes that were upregulated during physiological responses to carbon limitation (relative to limitation by N, S or P; Boer et al. 2003). The upregulation of these genes during carbon limitation in Boer et al. (2003) might suggest that they were upregulated by carbon-limited ancestral strains in F99 and J05. The 157 genes upregulated in response to carbon limitation in Boer et al. (2003) account for approximately 3% of predicted CDSs. Relative to this proportion, these genes were overrepresented among genes with higher expression in AN than SE strains in selection studies F99 and J05. Specifically, among these 157 genes, 11 were represented among the 69 genes upregulated in AN relative to SE strains in F99 (p<0.001, binomial test) and 21 were represented among the 63 genes upregulated in AN relative to SE strains in J05 (p<0.001, binomial test). We also found that these 21 and 11 genes encoded proteins that were carbon depleted relative to the rest of the proteome (J05: p=0.036; Mann–Whitney U-test, n=21, 5834; F99: p=0.026, Mann–Whitney U-test, n=11, 5844).
Finally, we examined carbon content biases in proteins whose genes show the opposite expression change after evolution than the genes we just studied. These genes have lower expression in ancestral (AN) strains than in selected (SE) strains. Their 178 and 62 gene products (as identified by Ferea et al. (1999) and Jansen et al. (2005), respectively) did not have carbon content significantly different from the rest of the proteome (J05: p=0.065, Mann–Whitney U-test, n=178, 5677; F99: p=0.144, Mann–Whitney U-test, n=62, 5793). These proteins were also not different from the rest of the proteome in their respiratory energy cost (J05: p=0.137, Mann–Whitney U-test, n=178, 5677; F99: p=0.405, Mann–Whitney U-test, n=62, 5793) or fermentative energy cost (J05: p=0.294, Mann–Whitney U-test, n=178, 5677; F99: p=0.062, Mann–Whitney U-test, n=62, 5793).
(c) Carbon assimilatory proteins show similar expression changes and cost biases
Baudouin-Cornu et al. (2001) identified a set of 21 yeast carbon assimilatory proteins that are present in our dataset. These proteins were significantly depleted in carbon relative to the rest of the proteome (p<0.001, Mann–Whitney U-test, n=21, 5834; Baudouin-Cornu et al. 2001). They also had significantly lower fermentative energy cost (p=0.001, Mann–Whitney U-test, n=21, 5834) but not respiratory energy cost (p=0.125, Mann–Whitney U-test, n=21, 5834), relative to the rest of the proteome. These carbon assimilatory proteins account for 5 (out of 63; Jansen et al. 2005) and 8 (out of 69; Ferea et al. 1999) proteins whose genes were expressed more highly in ancestral (AN) versus artificially selected (SE) strains. Excluding these assimilatory protein products, genes whose expression was higher in ancestral (AN) than artificially selected (SE) strains still had significantly lower carbon content relative to the rest of the proteome (J05: p=0.001, Mann–Whitney U-test, n=58, 5776; F99: p=0.021, Mann–Whitney U-test, n=61, 5773). Also, the assimilatory proteins that had greater expression in ancestral (AN) than artificially selected (SE) strains had lower carbon content than the remainder of the assimilatory proteins (J05: p=0.032, Mann–Whitney U-test, n=5, 16; F99: p=0.002, Mann–Whitney U-test, n=8, 13).
Only one of the assimilatory proteins identified by Baudouin-Cornu et al. (2001) had lower expression in ancestral (AN) than in selected (SE) strains, and in only one of the two artificial selection studies (F99).
(d) Duplication, expression, and energetic and elemental costs
Out of 5855 genes, 1502 yeast genes in our reference dataset have at least one duplicate. These genes tend to have higher expression levels (using CAI as a surrogate of expression; Sharp & Li 1987; Coghlan & Wolfe 2000) than genes that have no duplicates (p<0.001, Mann–Whitney U-test, n=1413, 4353 and Papp et al. 2003). Genes whose protein products have relatively low elemental and energetic costs also have more (surviving) duplicates. Specifically, genes with at least one duplicate had gene products with significantly lower carbon and nitrogen content than those with no duplicates (for both C and N, p<0.001, Mann–Whitney U-tests, n=1502, 4353; figure 3). There was no difference in the energetic costs of proteins whose genes had duplicates and those that had no duplicates (fermentative energy: p=0.762; respiratory energy, p=0.526; Mann–Whitney U-tests, n=1502, 4353).
Figure 3.
Mean (±2 s.e.) (a) carbon content and (b) nitrogen content (atoms per amino acid) of proteins with at least one duplicate, and proteins with no duplicates, in yeast.
A possible confounding factor in this analysis is gene expression, because genes with high expression can exhibit amino acid compositions consistent with adaptation for reduced elemental cost (e.g. sulphur, Fauchon et al. 2002; nitrogen, Elser et al. 2006) and energetic costs (e.g. Akashi & Gojobori 2002; Heizer et al. 2006). Indeed, we found that for yeast proteins, carbon content, nitrogen content and energetic cost (per amino acid) are each negatively associated with CAI (carbon: rs=−0.143, p<0.001; nitrogen: rs=−0.162, p<0.001; fermentative energy: rs=−0.277, p<0.001; respiratory energy: rs=−0.164, p<0.001; n=5766). These negative associations raise the possibility that high CAI values drive our observations of low carbon and nitrogen content for gene products of genes that have at least one duplicate.
We thus also needed to ask whether the association between gene duplication and low carbon or nitrogen content is independent of the influence of CAI. In order to do so, we fit linear regressions to the relationships between protein elemental costs and CAI (log transformed), and calculated standardized residuals. For both nitrogen and carbon, these residuals had a strong tendency to be lower for the gene products of genes with at least one duplicate than for genes with no duplicates (both N and C, p<0.001, Mann–Whitney U-tests, n=1413, 4353). In summary, high expression does not fully explain the low carbon and nitrogen content of proteins encoded by duplicate genes.
We next subdivided genes into different functional categories based on the MIPS Functional Catalogue (Ruepp et al. 2004). We did so to ask whether the different elemental contents of single copy and duplicated genes persisted in these categories. The answer is yes, with some exceptions. In other words, within functional categories, protein carbon content (table 1) and nitrogen content (see electronic supplementary material) were often higher for single copy genes than for duplicated genes.
Table 1.
Carbon content biases of proteins encoded by single-copy (S) and duplicated (D) genes in different protein functional categories. (Column 2 shows the number of analysed proteins, n, in each category, separately, for S and D genes. Column 3 shows the Spearman's rank correlation coefficients, rs (CAI), between carbon content per amino acid and CAI. Column 4 shows the difference in median carbon content between S and D genes. p-values in parentheses are derived from Mann–Whitney U-tests. Values in bold indicate p<0.01 for the difference between carbon cost of S and D genes. Note that duplicate genes always have lower C content, regardless of category. Column 5 shows p-values (P(r)) for Mann–Whitney U-tests of residuals from linear regressions between carbon content and (log) CAI, for all categories where the association between CAI and carbon content was significant. Differences here indicate that high expression (CAI) cannot fully explain carbon cost differences between S and D genes (*p<0.05, **p<0.01, ***p<0.001).)
| function | n (S, D) | rs (CAI) | median S−D (×10−2) | p | P(r) |
|---|---|---|---|---|---|
| metabolism or energy | 1050, 595a | −0.147*** | 3.10 | (<10−3) | 0.010 |
| cell cycle and DNA processing | 793, 200a | −0.064* | 4.96 | (<10−3) | 0.001 |
| transcription | 844, 178 | 0.025 | 2.92 | (0.023) | |
| translation | 330, 147 | −0.257*** | 4.90 | (0.006) | 0.516 |
| protein fate | 864, 281a | −0.077** | 4.31 | (0.001) | 0.007 |
| cellular transport, transport facilitation, transport routes | 703, 325 | −0.089** | 2.40 | (0.006) | 0.046 |
| cell rescue defence and virulence | 337, 215 | −0.227*** | 1.70 | (0.112) | 0.781 |
| biogenesis of cellular components | 659, 197 | −0.092** | 6.21 | (<10−3) | 0.041 |
| cell type differentiation | 301, 146 | 0.004 | 2.31 | (0.180) |
One duplicated CDS did not have a CAI value available (analyses of CAI have n−1 data points).
As mentioned above, 1502 yeast genes had at least one duplicate, accounting for about 25.7% of the genes in our reference dataset of 5855 gene products. Relative to this percentage, duplicate genes were over-represented among genes upregulated during glucose limitation. Specifically, out of 157 yeast genes that were upregulated during growth on low glucose (Boer et al. 2003), 62 (39.5%) had at least one duplicate (p<0.001, binomial test). Similarly, duplicate genes were over-represented among genes that were expressed more highly by ancestral (AN) than artificially selected (SE) strains: out of 63 genes that were expressed more by AN than SE in Jansen et al. (2005), 32 (50.8%) had at least one duplicate (binomial test, p<0.001), and out of 69 genes expressed more by AN than SE in Ferea et al. (1999), 28 (40.6%) had at least one duplicate (binomial test, p=0.005). This over-representation is consistent with the observations that (i) genes upregulated under glucose limitation and (ii) genes with duplicates tend to have carbon-depleted products.
4. Discussion
Our data show two ways in which protein nutrient costs interact with genome evolution. First, proteins whose genes are upregulated in carbon-limited (ancestral) yeast cells have low carbon costs, relative to strains that are artificially selected under low carbon availability. Second, genes with duplicates tend to have protein products with lower carbon and nitrogen content than singletons, which suggests that these costs influence the survival of duplicates.
During carbon limitation, genes more highly expressed in ancestral strains than in strains evolved under carbon limitation had carbon-poor protein products. This pattern suggests that carbon sparing occurs in ancestral (AN) strains, relative to selected (SE) strains. Selected strains may have acquired new adaptations to carbon limitation, leading to a reduced tendency to upregulate carbon-poor proteins that are part of an initial response to carbon limitation by ancestral strains (table 2; response 2). This interpretation is supported by the overlap between sets of genes that were upregulated in carbon-limited conditions in Boer et al. (2003; relative to N, S and P limitation) and those with higher expression in ancestral strains relative to artificially selected strains in Ferea et al. (1999) and Jansen et al. (2005). The proteins encoded by these overlapping gene sets tend to be carbon poor.
Table 2.
Two possible (hypothetical) responses by artificially selected and ancestral strains to carbon limitation and predictions for the carbon content of differentially expressed genes. (Our observations and hypotheses receiving some support are indicated in bold.)
| protein carbon content | ||
|---|---|---|
| selected strains' response to carbon limitation | gene expression level ancestral>selected | gene expression level ancestral<selected |
| (1) carbon sparing via expression changes | high carbon | low carbon |
| (2) relaxation of carbon sparing | ||
| ancestral strains' response to carbon limitation: | ||
| (a) carbon sparing via expression changes | low carbon | high carbon |
| (b) upregulation of proteins required during carbon limitation, which have low carbon content | low carbon | no bias in carbon content |
Several different types of adaptations in artificially selected strains could explain the relaxation of carbon sparing. One possibility is that the strains evolved greater glucose affinity (Dykhuizen & Hartl 1981; Helling et al. 1987; Jansen et al. 2005) and thus experienced partial relief from carbon-limitation conditions. Another possibility is that the carbon-poor proteins were upregulated as part of a carbon-limitation response in ancestral strains and that refinement of this response by natural selection reduced its magnitude in artificially selected strains.
One of the two main alternative hypotheses in our study was that laboratory selection on carbon limitation would lead to transcription-mediated carbon sparing in selected strains, relative to ancestral strains (table 2, response 1). We found no evidence consistent with this hypothesis. Instead, our results suggest that other evolutionary adaptations may modify the response to nutrient limitation, adaptations that decouple environmental nutrient limitations from protein composition. Such adaptations may help explain why no clear links have been found between the mean elemental composition of prokaryotic proteomes and habitat nutrient availability (Baudouin-Cornu et al. 2004; Bragg & Hyder 2004; Bragg et al. 2006).
During responses to carbon limitation, two non-exclusive adaptive responses could explain the upregulation of genes with carbon-poor proteins by ancestral strains. First, the cell may ‘attempt’ to save carbon by upregulating carbon-poor proteins and by downregulating carbon-rich proteins (table 2, response 2a). Second, the cell may upregulate proteins specifically useful to cope with the carbon limitation. These proteins may have evolved to have low carbon content, so they can be expressed more easily when carbon is scarce (table 2, response 2b). This reduction may take considerably longer than mere changes in gene expression levels. Our data are fully consistent with the latter scenario (response 2b) and only partially consistent with the former scenario (response 2a). This is because downregulated proteins were not carbon rich, which might be expected as part of an attempt to save carbon (response 2a), but not if natural selection had acted to reduce the carbon content of specific proteins that are important during carbon limitation (response 2b). We note that the available data do not allow us to rule out either scenario (response 2a or 2b) with certainty.
Overall, our observations are consistent with previous observations of carbon depletion in carbon assimilatory proteins, which has been attributed to selective reduction of carbon costs of these proteins, in order to help assimilatory pathways function during carbon shortages (Baudouin-Cornu et al. 2001). Indeed, some of the assimilatory proteins studied by Baudouin-Cornu et al. (2001) are encoded by genes that are upregulated during carbon limitation in ancestral strains. In other words, the upregulated and carbon-depleted gene products we identified include some of these assimilatory proteins. However, when we excluded these proteins, we still observed upregulation of carbon-depleted proteins. Thus, known assimilatory proteins form part of the response we see, but do not explain all of it.
Several studies have reported upregulation of sulphur-poor proteins (or their genes) during physiological responses to sulphur limitation (Cuhel et al. 1981; Mazel & Marlière 1989; Fauchon et al. 2002; Boer et al. 2003). We observed upregulation of genes encoding carbon-poor proteins by carbon-limited cells, but our observations are different from those for sulphur in an important way. We did not find low carbon content in proteins whose genes were upregulated by carbon-limited yeast, relative to yeast limited by other nutrients (N, S or P; from Boer et al. 2003). We detected carbon sparing only when expression by ancestral strains was compared to expression by strains that had been selected under carbon limitation (from Ferea et al. 1999; Jansen et al. 2005). A possible explanation is that ancestral strains have a carbon-sparing response, but it is weaker than for sulphur, or was less strongly elicited under the experimental conditions (in Boer et al. 2003). In other words, possibly carbon sparing is masked here by other differentially regulated genes.
A major factor influencing genome evolution is gene duplication (Ohno 1970; Lynch & Conery 2000; Rubin et al. 2000; Conant & Wagner 2002; Gu et al. 2002). In particular, retention and loss of gene duplicates may shape genomes over long evolutionary time-scales. The fate of duplicated genes is influenced not only by drift, but also by natural selection (Lynch et al. 2001), e.g. through the increased gene dosage and expression costs that are caused by gene duplications (Papp et al. 2003; Wagner 2005). Protein elemental costs potentially influence the retention of duplicates in several ways. A gene duplication may confer an advantage if one paralogue encodes a protein with a low requirement for an element and can be upregulated when that element is scarce (e.g. sulphur, Mazel & Marlière 1989; zinc, Panina et al. 2003). Here, we identify a possible additional role for carbon and nitrogen costs in evolution by gene duplication. Specifically, our analyses show that genes with at least one duplicate tend to have gene products with lower carbon and nitrogen content than genes with no duplicates. This suggests that duplicates of genes with high nitrogen or carbon content may be more likely to be eliminated from the genome by natural selection, due to their cost.
In summary, the initial response by yeast to carbon limitation may involve the upregulation of carbon-poor proteins. Importantly, this response is reduced after artificial selection under carbon limitation. Carbon and nitrogen costs of gene expression may influence the fate of duplicate genes, suggesting how nutrient availability could influence proteomic elemental composition in the long term. Taken together, our analyses help explain how the elemental composition of proteomes evolves.
Acknowledgments
We thank J. Brown, R. Charnov, E. Loker and J. Elser for helpful discussion. Comments from four anonymous referees greatly improved this manuscript. J.G.B. was supported by an NSF Biocomplexity grant (DEB-0083422).
Supplementary Material
Nitrogen content biases of proteins encoded by single-copy (S) and duplicated (D) genes in different protein functional categories
References
- Akashi H, Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilus. Proc. Natl Acad. Sci. USA. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. doi:10.1073/pnas.062526999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F, Madden T.L, Schaffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin-Cornu P, Surdin-Kerjan Y, Marlière P, Thomas D. Molecular evolution of protein atomic composition. Science. 2001;293:297–300. doi: 10.1126/science.1061052. doi:10.1126/science.1061052 [DOI] [PubMed] [Google Scholar]
- Baudouin-Cornu P, Schuerer K, Marlière P, Thomas D. Intimate evolution of proteins Proteome atomic content correlates with genome base composition. J. Biol. Chem. 2004;279:5421–5428. doi: 10.1074/jbc.M306415200. doi:10.1074/jbc.M306415200 [DOI] [PubMed] [Google Scholar]
- Boer V.M, de Winde J.H, Pronk J.T, Piper M.D.W. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus or sulfur. J. Biol. Chem. 2003;278:3265–3274. doi: 10.1074/jbc.M209759200. doi:10.1074/jbc.M209759200 [DOI] [PubMed] [Google Scholar]
- Bragg J.G, Hyder C.L. Nitrogen versus carbon use in prokaryotic genomes and proteomes. Proc. R. Soc. B. 2004;271(Suppl. 5):S374–S377. doi: 10.1098/rsbl.2004.0193. doi:10.1098/rsbl.2004.0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg J.G, Thomas D, Baudouin-Cornu P. Variation among species in proteomic sulphur content is related to environmental conditions. Proc. R. Soc. B. 2006;273:1293–1300. doi: 10.1098/rspb.2005.3441. doi:10.1098/rspb.2005.3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Wolfe K.H. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast. 2000;16:1131–1145. doi: 10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F. doi:10.1002/1097-0061(20000915)16:12<1131::AID-YEA609>3.0.CO;2-F [DOI] [PubMed] [Google Scholar]
- Conant G.C, Wagner A. GenomeHistory: a software tool and its application to fully sequenced genomes. Nucleic Acids Res. 2002;30:3378–3386. doi: 10.1093/nar/gkf449. doi:10.1093/nar/gkf449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuhel R.L, Taylor C.D, Jannasch H.W. Assimilatory sulfur metabolism in marine mircoorganisms: sulfur metabolism, growth and protein synthesis of Pseudomonas halodurans and Alteromonas luteo-violaceus during sulfate limitation. Arch. Microbiol. 1981;130:1–7. doi:10.1007/BF00527063 [Google Scholar]
- Dykhuizen D, Hartl D. Evolution of competitive ability in Escherichia coli. Evolution. 1981;35:581–594. doi: 10.1111/j.1558-5646.1981.tb04919.x. doi:10.2307/2408204 [DOI] [PubMed] [Google Scholar]
- Elser J.J, Fagan W.F, Subramanian S, Kumar S. Signatures of ecological resource availability in the animal and plant proteomes. Mol. Biol. Evol. 2006;23:1946–1951. doi: 10.1093/molbev/msl068. doi:10.1093/molbev/msl068 [DOI] [PubMed] [Google Scholar]
- Fauchon M, et al. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell. 2002;9:713–723. doi: 10.1016/s1097-2765(02)00500-2. doi:10.1016/S1097-2765(02)00500-2 [DOI] [PubMed] [Google Scholar]
- Ferea T.L, Botstein D, Brown P.O, Rosenzweig R.F. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl Acad. Sci. USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. doi:10.1073/pnas.96.17.9721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen F.-C, Bouman P, Li W.-H. Extent of gene duplication in the genomes of Drosophila, nematode and yeast. Mol. Biol. Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Heizer E.M, Jr, Raiford D.W, Raymer M.L, Doom T.E, Miller R.V, Krane D.E. Amino acid cost and codon-usage biases in 6 prokaryotic genomes: a whole-genome analysis. Mol. Biol. Evol. 2006;23:1670–1680. doi: 10.1093/molbev/msl029. doi:10.1093/molbev/msl029 [DOI] [PubMed] [Google Scholar]
- Helling R.B, Vargas C.N, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:549–558. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M.L.A, Diderich J.A, Mashego M, Hassane A, de Winde J.H, Daran-Lapujade P, Pronk J.T. Prolongled selection in aerobic, glucose-limited chemostat cultures of Saccharomyces cerevisiae causes a partial loss of glycolytic capacity. Microbiology. 2005;151:1657–1669. doi: 10.1099/mic.0.27577-0. doi:10.1099/mic.0.27577-0 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. doi:10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel D, Marlière P. Adaptive eradication of methionine and cysteine from cyanobacterial light-harvesting proteins. Nature. 1989;341:245–248. doi: 10.1038/341245a0. doi:10.1038/341245a0 [DOI] [PubMed] [Google Scholar]
- Ohno S. Springer; New York, NY: 1970. Evolution by gene duplication. [Google Scholar]
- Panina E.M, Mironov A.A, Gelfand M.S. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis and rearrangement of ribosomal proteins. Proc. Natl Acad. Sci. USA. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. doi:10.1073/pnas.1733691100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst L.D. Evolution of cis-regulatory elements in duplicated genes of yeast. Trends Genet. 2003;19:417–422. doi: 10.1016/S0168-9525(03)00174-4. doi:10.1016/S0168-9525(03)00174-4 [DOI] [PubMed] [Google Scholar]
- Pardee A.B. Purification and properties of a sulfate-binding protein from Salmonella typhimurium. J. Biol. Chem. 1966;241:5886–5892. [PubMed] [Google Scholar]
- Pascal G, Médigue C, Danchin A. Persistent biases in the amino acid composition of prokaryotic proteins. Bioessays. 2006;28:726–738. doi: 10.1002/bies.20431. doi:10.1002/bies.20431 [DOI] [PubMed] [Google Scholar]
- Richmond R.C. Non-Darwinian evolution: a critique. Nature. 1970;255:223–225. doi: 10.1038/2251025a0. [DOI] [PubMed] [Google Scholar]
- Rocha E.P.C, Sekowska A, Danchin A. Sulphur islands in the Escherichia coli genome: markers of the cell's architecture? FEBS Lett. 2000;476:8–11. doi: 10.1016/s0014-5793(00)01660-4. doi:10.1016/S0014-5793(00)01660-4 [DOI] [PubMed] [Google Scholar]
- Rubin G.M, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. doi:10.1126/science.287.5461.2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. doi:10.1093/nar/gkh894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M, Li W.-H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. doi:10.1093/nar/15.3.1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D, Higgins D.G, Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 2005;22:1365–1374. doi: 10.1093/molbev/msi126. doi:10.1093/molbev/msi126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nitrogen content biases of proteins encoded by single-copy (S) and duplicated (D) genes in different protein functional categories