Abstract
PC4 is a member of the proprotein convertase family of serine proteases implicated in the processing of a variety of polypeptides including prohormones, proneuropeptides, and cell surface proteins. In rodents, PC4 transcripts have been detected in spermatocytes and round spermatids exclusively, suggesting a reproductive function for this enzyme. In an effort to elucidate this function, we have disrupted its locus (Pcsk4) by homologous recombination in embryonic stem cells and have produced mice carrying the mutation. In intercrosses of heterozygous mutant mice, there was low transmission of the mutant Pcsk4 allele to the progeny, resulting in lower than expected incidence of heterozygosity and null homozygosity. The in vivo fertility of homozygous mutant males was severely impaired in the absence of any evident spermatogenic abnormality. In vitro, the fertilizing ability of Pcsk4 null spermatozoa was also found to be significantly reduced. Moreover, eggs fertilized by these spermatozoa failed to grow to the blastocyst stage. These results suggest that PC4 in the male may be important for achieving fertilization and for supporting early embryonic development in mice.
Keywords: gene inactivation, homologous recombination, allele transmission, sperm
Proprotein convertases (PCs) constitute a family of serine proteases structurally related to bacterial subtilisins and to yeast kexin. Seven eukaryotic members of this family are currently known. Their discovery and characterization have been described in recent reviews and articles (1–8). They are: PC1 (also known as PC3), PC2, furin, PC4, PC5 (also known as PC6), PACE4, and PC7 (also known as PC8 or LPC). The conserved homology of protein sequence and gene structure among these enzymes suggests that they evolved from a common ancestral gene through various mutagenic alterations, including duplications, insertions, deletions, and translocations (8, 9). Their loci (named Pcsk1–7) are distributed on different chromosomes in human and mouse, except for furin and PACE4, which map close to one another (8, 10).
PCs cleave precursor polypeptides at specific basic residues, most often after selected paired basic residues, to generate bioactive peptides and proteins. Included among their substrates are a wide variety of molecules such as prohormones, proneuropeptides, precursors to growth factors, cell surface receptors, and viral surface glycoproteins (1–5).
There are varying degrees of overlaps in the histological distribution and proteolytic activity of convertases, suggesting both redundancy and specificity in their biological functions (1, 2, 4, 5, 11–14).
Insight into the specific function and the physiological relevance of a convertase can be gained by studying a mutant animal model in which its gene is inactivated. No spontaneous phenotypic convertase mutant animal is currently available. As an alternative, induced mutant mice can be generated from chimera produced with embryonic stem (ES) cells in which Pcsk genes have been inactivated ex vivo by homologous recombination (15).
We have applied this technology to determine the biological relevance of the testis-specific convertase PC4. Messenger RNA for this convertase has been detected unambiguously only in spermatocytes and round spermatids of rat (16) and in round spermatids of mouse (17, 18). We have disrupted its locus (Pcsk4) in mouse ES cells and have used these cells to generate mice lacking PC4. Consistent with the pattern of expression of this enzyme, male Pcsk4 mutant mice exhibit significantly reduced fertility.
MATERIALS AND METHODS
Construction of the Pcsk4 Gene Disruption Vector.
An 11-kb DNA fragment containing the entire Pcsk4 gene was cloned from a 129/SvJ mouse genomic library in λFixII (Stratagene) using a 0.2-kb DNA fragment representing the first intron of the corresponding BALB/c gene (19). The homologous recombination vector was constructed into the backbone of the promoter trap plasmid pSAβgal (20), which carries a lacZ gene preceded by an acceptor splice site and followed by the bacterial neomycin phosphotransferase gene (neoR) under the phosphoglycerate kinase (PGK) promoter. A 2.3-kb BstZ–SpeI fragment extending from the promoter to the second intron of the Pcsk4 gene was inserted into pSAβgal cut with NotI and SpeI, upstream to the lacZ gene. Then a 2.6-kb SalI–XhoI Pcsk4 gene fragment extending from intron 6 to the middle of the exon 13 was inserted into homonymous sites after the neoR gene. Finally, an XhoI–SalI fragment containing the herpes simplex virus thymidine kinase (tk) gene under the PGK promoter was introduced into the XhoI site at the 3′ end of the Pcsk4 gene insert to generate the final construct called pPcsk4/SAβgalTK.
Production of Pcsk4 Null Mice.
The Pcsk4/SAβgalTK insert was excised from the construct and electroporated into E14TG2a ES cells (21). Clones were selected for resistance to the cytotoxic effects of 150 μg/ml G418 (neoR gene integration) and 0.25 μM 1-(2-fluoro-β-d-arabinofuranosyl)-5-iodouracyl (loss of the tk gene). They were screened by Southern blot analysis for integration of the lacZ and PGKneoR into the Pcsk4 locus, using probes flanking either end of the homology region. Five homologous recombinant clones were injected into the blastocoel cavity of C57BL/6J (B6) blastocysts. These were implanted into pseudopregnant mice. Male chimera born from these foster mothers were crossed to B6 females to generate heterozygous Pcsk4 mutant mice. These were subsequently intercrossed to homozygosity (22).
Blot Analyses.
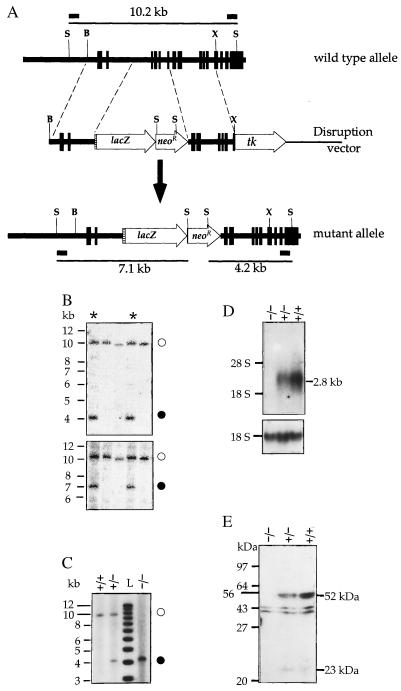
Southern blot analysis for homologous integration of the disrupting sequences was conducted on SphI digests of genomic DNA. This enzyme cleaves the Pcsk4 gene outside the homology region used in the pPcsk4/SAβgalTK construct, producing a 10.2-kb fragment that extends from the promoter to exon 15. Two Pcsk4 gene fragments were used as probes: a 0.43-kb BstZI–XbaI fragment and a 0.56-kb BstZI–SphI fragment, located 5′ and 3′ to the homology region, respectively. Both probes recognized a 10.2-kb fragment of the wild-type allele, but should detect shorter fragments (of 7.1 kb and 4.2 kb with the 5′ and 3′ probe, respectively) in the recombined allele due to the presence of SphI sites in the lacZ–PGKneoR insertion (see Fig. 1A). DNA probes were radiolabeled by the multiple priming method. The probe for Northern blot analysis was a radiolabeled cRNA transcribed from a 1.7-kb rat PC4 cDNA fragment covering exons 7–15.
Figure 1.
(A) Disruption of the Pcsk4 gene. Vertical boxes represent exons, the hatched area an acceptor splice site, and short horizontal bars DNA probe locations. B, BstZI; X, XhoI; S, SphI. (B and C) Southern blot genotyping of ES cell clones (B) and mouse tail tips (C) with a 3′ (B Upper and C) or the 5′ probe (B Lower). Open circles, solid circles, and asterisks identify the wild-type allele, the mutant allele, and recombinant clones, respectively. L stands for 1-kb ladder. (D) Northern blot analysis of testicular RNA of wild-type (+/+), heterozygous mutant (−/+), and homozygous mutant (−/−) mice for PC4 mRNA (Upper) and 18S rRNA (Lower). (E) Western blot analysis of epididymal sperm. The two immunoreactive proteins of about 52 and 23 kDa detectable in wild-type and heterozygote mouse samples are PC4 related. They were not observed after the antiserum had been preabsorbed with purified PC4 (not shown), unlike the two proteins of about 40 and 44 kDa detectable in all three samples, which are likely due to a nonspecific immunoreaction.
For Western blot analysis, epididymal sperm cells were washed by three rounds of sedimentation at 600 × g and resuspension in 1 ml of phosphate buffered saline. They were then lysed in a buffer made of 50 mM Tris⋅HCl (pH 7.5), 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 1 μg/ml pepstatin, and 2 μg/ml aprotinin; after a brief sonication on ice, the homogenates were centrifuged for 10 min at 10,000 × g and 4°C; supernatants were collected and their protein concentration determined. Proteins (20 μg) were separated by SDS/PAGE (10% gel) and analyzed by chemiluminescence Western blot analysis (23) using a rabbit antiserum against mouse PC4 (dilution 1:1,000).
Histology.
Testes were decapsulated, fixed in Bouin’s solution, cut into 25-mm sections that were stained for toluidine blue, and microscopically examined. For lacZ expression analysis, testes or testicular sections were stained for β-galactosidase (β-gal) activity using the chromogenic substrate 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (24).
Sperm Motility Analysis.
Spermatozoa were collected from cauda epididymis in fertilization medium and capacitated for 60 min at 37°C in 5% CO2/5% O2/90% air. Motility parameters were measured using the Cell Soft (Cryo Resources, Montgomery, NY) computer-assisted digital analysis system as described by Kodama et al. (25). The following parameters were measured: velocity (curvilinear velocity, μm/sec, the total distance divided by time), linearity (the distance in straight line divided by actual distance), amplitude of lateral head displacement (ALH, μm, the deviation of the head from the mean head trajectory), ALHave (the average of all the ALH measured), ALHmax (the maximal ALH observed for each spermatozoon), beat cross frequency (beats per second, the number of times that the spermatozoon head crosses the mean head trajectory per second), percent of circular cells (number of spermatozoa swimming in circles divided by the total number of motile spermatozoa), hyperactivation (the number of spermatozoa with a velocity > 136 μm/s, a linearity <5.5 and a ALHave >8 μm divided by the total number of motile spermatozoa).
In Vitro Fertilization (IVF).
IVF was performed as described by Kuribayashi and Gagnon (26). Briefly, mouse cauda epididymal spermatozoa capacitated in vitro were incubated with oocytes recovered from superovulated 6- to 8-week-old B6 wild-type female mice. Embryo development was followed by daily microscopic observation.
RESULTS AND DISCUSSION
The Pcsk4 allele was disrupted in mouse ES cells by homologous recombination using the targeting vector shown in Fig. 1A. In this vector, the lacZ–neoR domain was flanked by two consecutive but noncontiguous DNA fragments of the Pcsk4 gene. The 5′ (2.3 kb) and the 3′ (2.6 kb) fragments are 1.9 kb apart in the normal gene (Fig. 1A). The intervening region extends from intron 2 to intron 6 and contains exons specifying two of the four residues of the catalytic domain of the PC4 enzyme (exon 4 for Asp and exon 6 for His). Integration of the lacZ–neoR domain into the Pcsk4 locus will delete exons 3–6, rendering enzymatically inactive any truncated PC4-related proteins that might be specified by possible alternate transcripts. In the recombined gene (Fig. 1A), the lacZ reporter gene was placed under the control of the Pcsk4 promoter. Transcription from this promoter will produce a fusion mRNA containing sequences derived from Pcsk4 exons 1 and 2 as well as from the lacZ gene, which will constitute a third exon. In this transcript, however, the lacZ open reading frame will be out of frame with the open reading frame specified by the two Pcsk4 exons. Biosynthesis of β-gal from the fusion mRNA will begin at an AUG near the 5′ end of the lacZ exon.
The construct was transfected into ES cells and clones were doubly selected for neoR integration and tk loss. Genomic DNA from these clones was digested with SphI and analyzed by Southern blot for homologous recombination using a 3′ probe. Of 248 ES clones thus screened, 9 showed the 10.2- and 4.2-kb SphI fragments diagnostic of lacZ–neoR integration in one Pcsk4 allele (Fig. 1B Upper) for a homologous recombination frequency of 3.6%. The 10.2- and 7.1-kb SphI fragments hybridizing to the 5′ probe were also observed in these nine clones (Fig. 1B Lower). This excluded any gross rearrangement at both ends of the homology region during recombination. Southern blot analysis with a neoR and a tk probe confirmed the presence of a single integration site at the Pcsk4 locus and the absence of any silent tk integration elsewhere in the genome of the selected clones (not shown).
Six recombinant ES cell clones were used to generate chimeric mice. When these chimeras were crossed to B6 mice, there was germ-line transmission of the disrupted allele from three of the clones. Only male chimeras transmitted the mutation. Mice derived from two independent ES cell clones (lines 417 and 420) were used for subsequent studies. Pcsk4−/+ F1 heterozygous animals were intercrossed to produce Pcsk4−/− F2 homozygotes. Screening for the mutation in the progeny was carried out by Southern blot analysis with the 3′ probe (Fig. 1C) or by PCR (not shown). Pcsk4−/− mice expressed no PC4 mRNA or protein in the testis, whereas, relative to their normal littermates, heterozygotes expressed reduced levels of these gene products (Fig. 1 D and E). Pcsk4 mutant mice, irrespective of gender, exhibited no obvious physical, growth, or behavioral abnormality.
Interestingly, the progeny of F1 heterozygote intercrosses consistently showed a lower than expected incidence of the Pcsk4+/− and Pcsk4−/− genotypes and a higher than expected incidence of the Pcsk4+/+ one. This transmission distortion was statistically significant for both the 417 and the 420 lines. Overall, of 632 F2 mice, 43.2% were Pcsk4+/+, 38.4% were Pcsk4+/−, and 18.4% were Pcsk4−/− (Table 1). These values represented significant deviations (P < 0.0001) from the 1:2:1 ratio expected from such heterozygote intercrosses if there were random allele segregation and inheritance. They point to the possibility of a reduced transmission of the mutant allele. This low transmission was also observed after genotyping 3.5-day embryos from 5 heterozygote matings in the 417 mutant line: of 92 embryos, 55 (60%) were +/+, 23 (25%) were +/−, and 14 (15%) were −/− (P < 0.0001), suggesting that fewer mutant gametes took part in fertilization or that fewer mutant preimplantation embryos survived.
Table 1.
Pcsk4 genotypes of the F2 progeny from F1 heterozygote intercrosses
| Genotype | No. of F2 mice per genotype
|
|||
|---|---|---|---|---|
| Line 417 (n = 260)
|
Line 420 (n = 372)
|
|||
| Observed | Expected | Observed | Expected | |
| +/+ | 130 (50.0) | 65 (25) | 143 (39.2) | 93 (25) |
| −/+ | 92 (35.4) | 130 (50) | 151 (40.6) | 186 (50) |
| −/− | 38 (14.6) | 65 (25) | 78 (21.0) | 93 (25) |
Lines 417 and 420 were derived from independent targeted ES clones. The number of genotyped mice in each line is indicated. The percent of each genotype is given in parentheses. The χ2 test was used to test significance. The frequency of the three genotypes in each line significantly deviates from that expected from Mendelian allele inheritance. P < 0.0001.
The consequence of Pcsk4 gene disruption on fertility was compared between heterozygote intercrosses and reciprocal crosses of heterozygous mice to homozygous targeted mice. In this evaluation, only F1 heterozygous mice were used because they were genetically identical. Homozygous targeted mice were of the F2 generation born of these heterozygotes. The number of productive matings and the average first litter size of all matings are shown in Table 2. All −/+ mice (controls) were fertile and their average litter size was 6.9 ± 1.8. Ten of the 13 matings of −/− females to −/+ males were productive and the average litter size was 2.4 ± 2.2 for all matings and 3.1 ± 1.6 among productive matings. Six of the 24 matings of −/+ females to −/− males were productive and the average litter size was 0.8 ± 1.7 for all matings and 3.3 ± 0.8 among productive matings. The decrease in fertility rate (percent of productive matings) was statistically far more significant for male homozygous mutant mice (P < 0.0001) than for female ones (P < 0.03), whereas the reduction in average litter size was of statistically comparable significance between genders (P < 0.005 for females and P < 0.0001 for males). Clearly, the combined effect of low rate of productive matings and small average litter size makes the mutation very deleterious for male fertility.
Table 2.
Reduced fertility of Pcsk4−/− mice
| Parents
|
n | Average litter size | % of productive matings |
|---|---|---|---|
| F M | |||
| −/+ × −/+ | 29 | 6.9 ± 1.8 | 100 |
| −/− × −/+ | 13 | 2.4 ± 2.2 (P < 0.005) | 77 (P < 0.03) |
| −/+ × −/− | 24 | 0.8 ± 1.7 (P < 0.0001) | 25 (P < 0.0001) |
Eight- to 12-week-old mice were mated for 4–6 weeks and the number of live births in the first litter was recorded. F, female; M, male; n, number of mating pairs. Values for average litter size represent means ± SD for all the mating pairs (n) in each genotype combination. Significance values (P) were calculated by Student’s t test for average litter sizes and by χ2 test for fractions of productive matings, using the −/+ matings as controls.
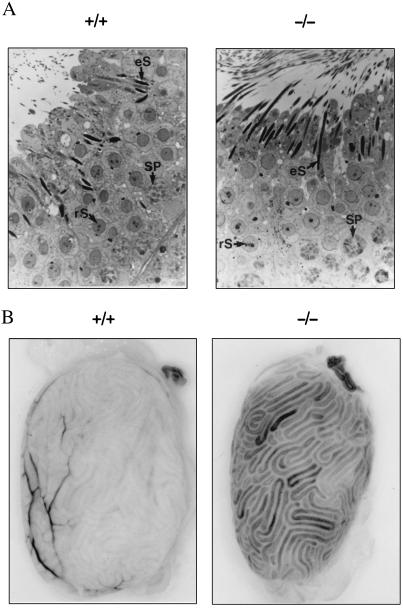
To determine if the reduced fertility of PC4-null male mice was due to a spermatogenic defect, a light microscopic analysis of the seminiferous epithelium was conducted on tubular sections showing various stages of the spermatogenic cycle. The histological characteristics of the epithelium, as well as the relative numbers of normal and degenerating cells, i.e., spermatogonia, spermatocytes, round and elongated spermatids in PC4-null mouse testes, were similar to that of normal littermates (Fig. 2A). Most motility parameters of PC4-null spermatozoa were also similar to those of wild-type mice, except for the percentage of hyperactivated spermatozoa which was lower (Table 3). Hyperactivation is a qualitative change in sperm motility that is thought to facilitate sperm penetration of oocyte vestments, a prerequisite to fertilization. Thus, PC4-null spermatozoa may be less efficient at oocyte penetration.
Figure 2.
(A) Toluidine blue stained semi-thin sections (×1,000) of seminiferous tubes from normal control (+/+) and Pcsk4 null mice (−/−). Arrows point at some pachytene spermatocytes (SP), round (rS) and elongated (eS) spermatids. (B) Staining for β-gal activity in the testis of wild-type (+/+) and mutant (−/−) mice. The reaction dark product is associated with testicular tubules.
Table 3.
Sperm motility analysis
| Parameter | Genotype
|
|
|---|---|---|
| +/+ | −/− | |
| Velocity (μm/s) | 170 ± 18 | 170 ± 19 |
| Linearity | 4.7 ± 0.1 | 5.0 ± 0.8 |
| ALHmax (μm) | 8.6 ± 1.4 | 7.8 ± 0.8 |
| ALHave (μm) | 7.4 ± 1.2 | 6.5 ± 0.6 |
| BCF (beat/s) | 12.4 ± 0.8 | 13.1 ± 0.9 |
| Circular cells | 4.5 ± 1.5 | 3.8 ± 1.2 |
| HA (%) | 12.9 ± 2.1 | 6.8 ± 1.4* |
Four mice of each genotype were studied. The number of spermatozoa analyzed range from 200 to 300 per mouse. The parameters examined are defined in Materials and Methods. Values represent the mean and SD of determinations on the four mice of each genotype.
PC4 null sperm undergo hyperactivation to a significantly lesser extent (P < 0.05).
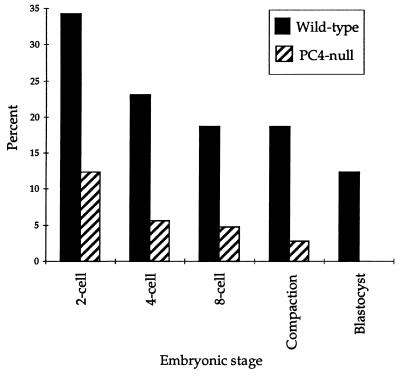
To further analyze the mutation effect on sperm competence for fertilization, as well as for its capacity to support early embryonic development, we conducted IVF experiments with spermatozoa collected from +/+ or −/− mice in our colony and oocytes collected from +/+ B6 mice. The rates of fertilization or development to the blastocyst stage were examined. With wild-type spermatozoa, 34% of the oocytes were fertilized, as measured by the formation of two cell embryos, 19% developed to the eight cell stage, and 12% to the blastocyst stage. With PC4-null spermatozoa, the values were 12%, 5%, and 0%, respectively (P < 0.0001) (Fig. 3), suggesting that PC4 deficiency causes reduced fertilization as well as early embryonic death.
Figure 3.
In vitro fertilizing ability of spermatozoa and their capacity to support embryo development. Spermatozoa from 4 wild-type and 4 PC4-null mice were studied in 4 separate experiments involving 283 and 250 wild-type B6 oocytes, respectively. Significance was determined by χ2 test. The differences between the two genotypes were significant in all cases (P < 0.0001).
Whether the lack of PC4 also causes lethality of postimplantation embryos in vivo has not been examined in this study. Such lethality could imply that PC4 may be transiently produced during embryonic development. Because the Pcsk4 mutant mouse we have generated carries the lacZ reporter gene under the control of the Pcsk4 promoter, verifying β-gal activity in embryos could be a rapid way of identifying potential sites of PC4 expression during development. We have tested e3.5 to e17 mutant embryos for β-gal activity and none tested positive. In contrast, testes of adult mutant mice contained this enzymatic activity (Fig. 2B). The β-gal reaction product was observed in the lumen of testicular tubules as expected, suggesting that the normal pattern of Pcsk4 promoter activation may have been preserved after disruption of the structural gene. Interestingly, in the female, a β-gal staining was observed in steroid-producing thecal–interstitial and luteal cells of the ovary (not shown). It will be warranted to determine whether an impairment of the steroidogenic function of these cells could be a cause of the reduced fertility PC4-null female mice.
The molecular basis of the fertility impairment caused by PC4 deficiency remains to be clarified. Serine proteases have been implicated in sperm maturation and capacitation as well as in fertilization. One of them, acrosin, is a major constituent of sperm acrosome. However, inactivation of its gene was found to have no deleterious effect on fertility (27). Our data suggest that PC4 could be important for reproduction. Several precursor polypeptides that need processing by convertase-like enzymes are expressed in the same testicular germ cells as PC4, among them precursors for enkephalins (28), for pituitary adenylate cyclase-activating peptide (29), for growth hormone-releasing hormone-related peptide (30), for nerve growth factor (31), and for fertilins α and β (32, 33). Of these proteins, only fertilins have been shown to be relevant for reproduction. These type 1 transmembrane glycoproteins are produced as large precursors in spermatocytes and spermatids; their processed forms are located on epididymal sperm head as αβ heterodimers (34). These complexes apparently participate in sperm–egg fusion by attaching to α6β1 integrins on oolemma (35). The possible failure by PC4-deficient spermatozoa to properly process such proteins may render them relatively incompetent for fertilization. To clarify the role of PC4 in reproduction and development, its natural substrates will need to be identified and their specific functions determined. A comparison of the sperm protein profiles between wild-type and Pcsk4 mutant mice should facilitate the identification of these substrates.
Acknowledgments
We thank K. A. Johnson for the pJAX40 vector, B. H. Koller and O. Smithies for the E14TG2a ES cells, D. S. Coopersmith for mouse colony management, J.-W. Van de Loo and W. J. M. Van de Ven for the anti-PC4 antiserum, and E. H. Leiter and L. Kozak for critical review of this manuscript. This work was supported by Medical Research Council of Canada grants to M.M., N.G.S., C.G., and M.C. and by a National Institutes of Health grant to The Jackson Laboratory.
ABBREVIATIONS
- ES
embryonic stem
- β-gal
β-galactosidase
- IVF
in vitro fertilization
- neoR
neomycin phosphotransferase gene
- PGK
phosphoglycerate kinase
- tk
thymidine kinase gene
- PCs
proprotein convertases
- ALH
amplitude of lateral head displacement
References
- 1.Van de Ven W J, Roebroek A J, Van Duijnhoven H L. Crit Rev Oncog. 1993;4:115–136. [PubMed] [Google Scholar]
- 2.Bresnahan P A, Hayflick J S, Molloy S S, Thomas G. In: Endoproteolysis of Growth Factors and Other Endocrine Precursor Proteins. Peng Loh Y, editor. Boca Raton, FL: CRC; 1993. pp. 225–250. [Google Scholar]
- 3.Seidah N G, Chrétien M. Methods Enzymol. 1994;244:175–188. doi: 10.1016/0076-6879(94)44015-8. [DOI] [PubMed] [Google Scholar]
- 4.Chrétien M, Mbikay M, Gaspar L, Seidah N G. Proc Assoc Am Physicians. 1995;107:47–66. [PubMed] [Google Scholar]
- 5.Rouille Y, Duguay S J, Lund K, Furuta M, Gong Q, Lipkind G, Oliva A A, Jr, Chan S J, Steiner D F. Front Neuroendocrinol. 1995;16:322–361. doi: 10.1006/frne.1995.1012. [DOI] [PubMed] [Google Scholar]
- 6.Bruzzaniti A, Goodge K, Jay P, Taviaux S A, Lam M H, Berta P, Martin T J, Moseley J M, Gillespie M T. Biochem J. 1996;314:727–731. doi: 10.1042/bj3140727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meerabux J, Yaspo M L, Roebroek A J, Van de Ven W J, Lister T A, Young B D. Cancer Res. 1996;56:448–451. [PubMed] [Google Scholar]
- 8.Seidah N G, Hamelin J, Mamarbachi M, Dong W, Tadros H, Mbikay M, Chrétien M, Day R. Proc Natl Acad Sci USA. 1996;93:3388–3393. doi: 10.1073/pnas.93.8.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouille Y, Martin S, Steiner D F. J Biol Chem. 1995;270:26488–26496. doi: 10.1074/jbc.270.44.26488. [DOI] [PubMed] [Google Scholar]
- 10.Mbikay M, Seidah N G, Chrétien M, Simpson E M. Genomics. 1995;26:123–129. doi: 10.1016/0888-7543(95)80090-9. [DOI] [PubMed] [Google Scholar]
- 11.Day R, Schafer M K, Watson S J, Chrétien M, Seidah N G. Mol Neuroendocrinol. 1992;6:485–497. doi: 10.1210/mend.6.3.1316544. [DOI] [PubMed] [Google Scholar]
- 12.Marcinkiewicz M, Ramla D, Seidah N G, Chrétien M. Endocrinology. 1994;135:1651–1660. doi: 10.1210/endo.135.4.7925129. [DOI] [PubMed] [Google Scholar]
- 13.Seidah N G, Chrétien M, Day R. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 14.Dong W, Marcinkiewicz M, Vieau D, Chrétien M, Seidah N G, Day R. J Neurosci. 1995;15:1778–1796. doi: 10.1523/JNEUROSCI.15-03-01778.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koller B H, Smithies O. Annu Rev Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 16.Seidah N G, Day R, Hamelin J, Gaspar A, Collard M W, Chrétien M. Mol Neuroendocrinol. 1992;6:1559–1570. doi: 10.1210/mend.6.10.1448111. [DOI] [PubMed] [Google Scholar]
- 17.Nakayama K, Kim W S, Torii S, Hosaka M, Nakagawa T, Ikemizu J, Baba T, Murakami K. J Biol Chem. 1992;267:5897–5900. [PubMed] [Google Scholar]
- 18.Torii S, Yamagishi T, Murakami K, Nakayama K. FEBS Lett. 1993;316:12–16. doi: 10.1016/0014-5793(93)81726-g. [DOI] [PubMed] [Google Scholar]
- 19.Mbikay M, Raffin-Sanson M L, Tadros H, Sirois F, Seidah N G, Chrétien M. Genomics. 1994;20:231–237. doi: 10.1006/geno.1994.1158. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 21.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature (London) 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 22.Joyner A L. Gene Targeting: A Practical Approach. Oxford: Oxford Univ. Press; 1995. [Google Scholar]
- 23.Linard C G, Tadros H, Sirois F, Mbikay M. Mol Cell Biochem. 1995;151:39–47. doi: 10.1007/BF01076894. [DOI] [PubMed] [Google Scholar]
- 24.Bonnerot C, Nicolas J F. Methods Enzymol. 1993;225:451–469. doi: 10.1016/0076-6879(93)25031-v. [DOI] [PubMed] [Google Scholar]
- 25.Kodama H, Kuribayashi Y, Gagnon C. J Androl. 1996;17:151–157. [PubMed] [Google Scholar]
- 26.Kuribayashi Y, Gagnon C. Fertil Steril. 1996;66:1012–1017. doi: 10.1016/s0015-0282(16)58699-3. [DOI] [PubMed] [Google Scholar]
- 27.Baba T, Azuma S, Kashiwabara S, Toyoda Y. J Biol Chem. 1994;269:31845–31849. [PubMed] [Google Scholar]
- 28.Kew D, Muffly K E, Kilpatrick D L. Proc Natl Acad Sci USA. 1990;87:9143–9147. doi: 10.1073/pnas.87.23.9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannibal J, Fahrenkrug J. Regul Pept. 1995;55:111–115. doi: 10.1016/0167-0115(94)00110-j. [DOI] [PubMed] [Google Scholar]
- 30.Breyer P R, Rothrock J K, Beaudry N, Pescovitz O H. Endocrinology. 1996;137:2159–2162. doi: 10.1210/endo.137.5.8612561. [DOI] [PubMed] [Google Scholar]
- 31.Ayer-LeLievre C, Olson L, Ebendal T, Hallbook F, Persson H. Proc Natl Acad Sci USA. 1988;85:2628–2632. doi: 10.1073/pnas.85.8.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blobel C P, Myles D G, Primakoff P, White J M. J Cell Biol. 1990;111:69–78. doi: 10.1083/jcb.111.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry A C, Gichuhi P M, Jones R, Hall L. Biochem J. 1995;307:843–850. doi: 10.1042/bj3070843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll D J, Dikegoros E, Koppel D E, Cowan A E. Dev Biol. 1995;168:429–437. doi: 10.1006/dbio.1995.1092. [DOI] [PubMed] [Google Scholar]
- 35.Almeida E A, Huovila A P, Sutherland A E, Stephens L E, Calarco P G, Shaw L M, Mercurio A M, Sonnenberg A, Primakoff P, Myles D G, White J M. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]