Abstract
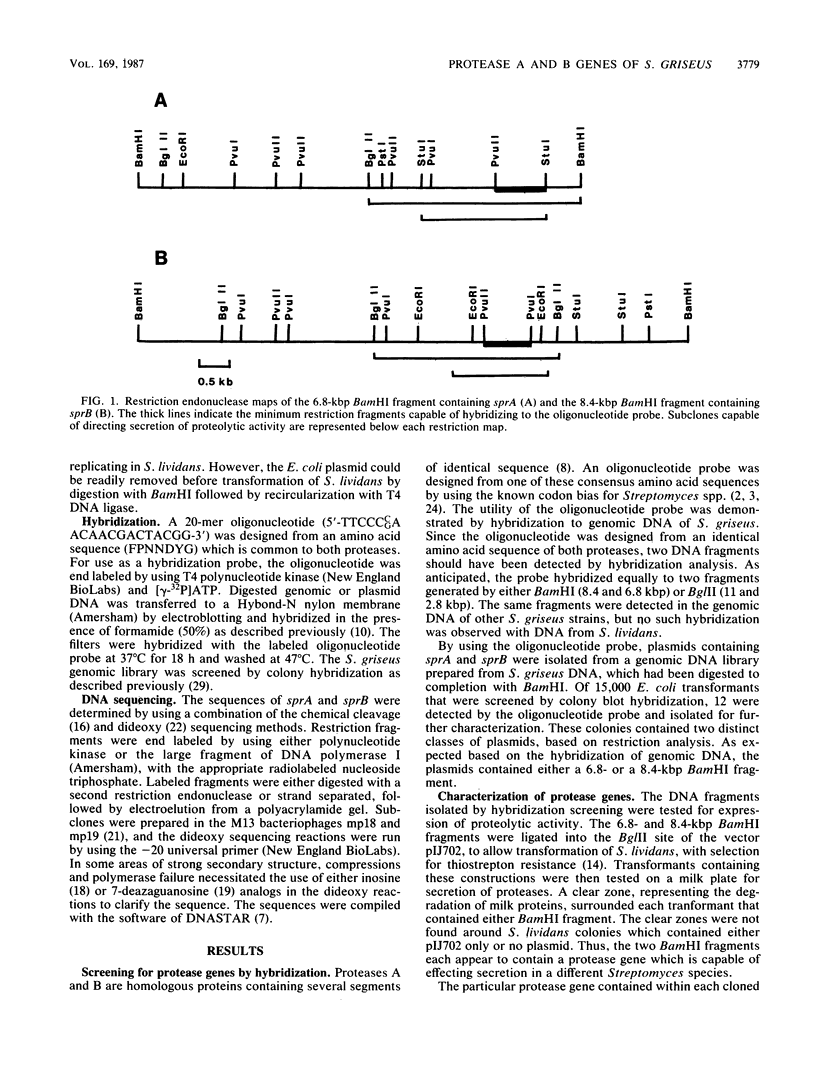
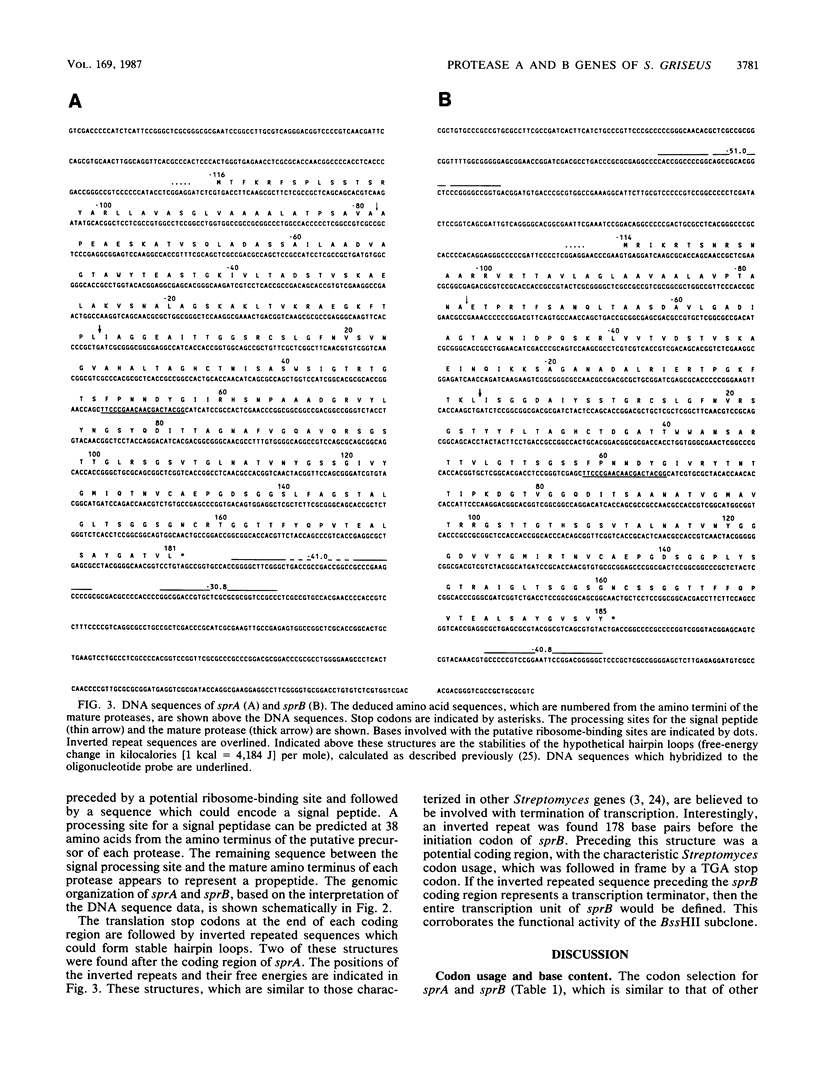
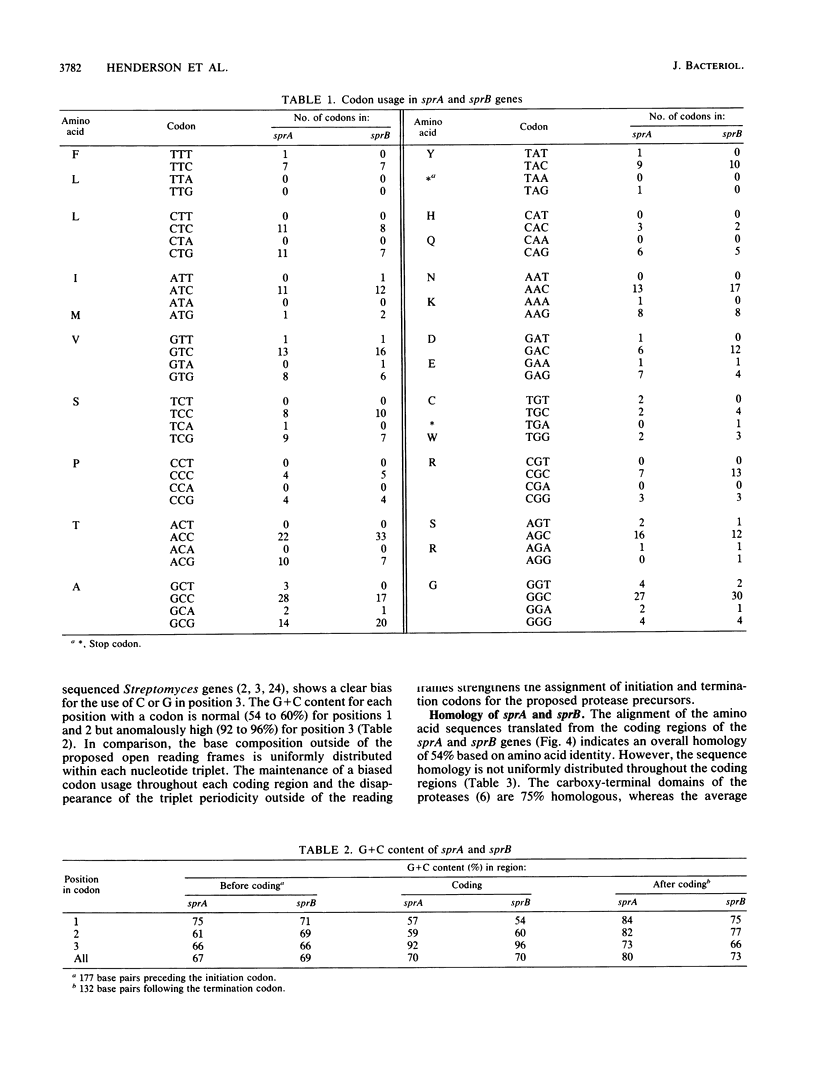
Protease A and protease B are extracellular proteins which are secreted by Streptomyces griseus. The genes encoding protease A (sprA) and protease B (sprB) were isolated from an S. griseus genomic library by using a synthetic oligonucleotide probe. Fragments containing sprA and sprB were characterized by hybridization and demonstration of proteolytic activity in Streptomyces lividans. Each DNA sequence contains a large open reading frame with the coding region of the mature protease situated at its carboxy terminus. The amino terminus of each reading frame appears to encode a 38-amino-acid signal peptide followed by a 76- or 78-amino-acid polypeptide, a propeptide, which is joined to the mature protease. Strong homology between the coding regions of the protease genes suggests that sprA and sprB originated by gene duplication.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahmsén L., Moks T., Nilsson B., Hellman U., Uhlén M. Analysis of signals for secretion in the staphylococcal protein A gene. EMBO J. 1985 Dec 30;4(13B):3901–3906. doi: 10.1002/j.1460-2075.1985.tb04164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernan V., Filpula D., Herber W., Bibb M., Katz E. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene. 1985;37(1-3):101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Bibb M., Schottel J. L., Cohen S. N. A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces. Nature. 1980 Apr 10;284(5756):526–531. doi: 10.1038/284526a0. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T., Thompson C. J. Gene cloning in Streptomyces. Curr Top Microbiol Immunol. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Delbaere L. T., Hutcheon W. L., James M. N., Thiessen W. E. Tertiary structural differences between microbial serine proteases and pancreatic serine enzymes. Nature. 1975 Oct 30;257(5529):758–763. doi: 10.1038/257758a0. [DOI] [PubMed] [Google Scholar]
- Doggett P. E., Blattner F. R. Personal access to sequence databases on personal computers. Nucleic Acids Res. 1986 Jan 10;14(1):611–619. doi: 10.1093/nar/14.1.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga M., Delbaere L. T., Brayer G. D., James M. N. Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure. J Mol Biol. 1985 Aug 5;184(3):479–502. doi: 10.1016/0022-2836(85)90296-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- James M. N., Sielecki A. R., Brayer G. D., Delbaere L. T., Bauer C. A. Structures of product and inhibitor complexes of Streptomyces griseus protease A at 1.8 A resolution. A model for serine protease catalysis. J Mol Biol. 1980 Nov 25;144(1):43–88. doi: 10.1016/0022-2836(80)90214-4. [DOI] [PubMed] [Google Scholar]
- Johnson P., Smillie L. B. The disulfide bridge sequences of a serine protease of wide specificity from Streptomyces griseus. Can J Biochem. 1971 May;49(5):548–562. doi: 10.1139/o71-082. [DOI] [PubMed] [Google Scholar]
- Jurásek J., Johnson P., Olafson R. W., Smillie L. B. An improved fractionation system for pronase on CM-sephadex. Can J Biochem. 1971 Nov;49(11):1195–1201. doi: 10.1139/o71-171. [DOI] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narahashi Y., Yoda K. Studies on proteolytic enzymes (pronase) of Streptomyces griseus K-1. 3. Purification and proteolytic specificity of alkaline proteinase C from pronase. J Biochem. 1973 Apr;73(4):831–841. doi: 10.1093/oxfordjournals.jbchem.a130146. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens G. J., Günzler W. A., Otting F., Frankus E., Flohé L. The complete amino acid sequence of low molecular mass urokinase from human urine. Hoppe Seylers Z Physiol Chem. 1982 Sep;363(9):1043–1058. doi: 10.1515/bchm2.1982.363.2.1043. [DOI] [PubMed] [Google Scholar]
- Thompson C. J., Gray G. S. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5190–5194. doi: 10.1073/pnas.80.17.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


