Abstract
In contrast to the distal half of the long arm of chromosome 21, the proximal half of approximately 20 megabases of DNA, including 21q11–21 bands, is low in GC content, CpG islands, and identified genes. Despite intensive searches, very few genes and cDNAs have been found in this region. Since the 21q11–21 region is associated with certain Down syndrome pathologies like mental retardation, the identification of relevant genes in this region is important. We used a different approach by constructing microdissection libraries specifically for this region and isolating unique sequence microclones for detailed molecular analysis. We found that this region is enriched with middle and low-copy repetitive sequences, and is also heavily methylated. By sequencing and homology analysis, we identified a significant number of genes/cDNAs, most of which appear to belong to gene families. In addition, we used unique sequence microclones in direct screening of cDNA libraries and isolated 12 cDNAs for this region. Thus, although the 21q11–21 region is gene poor, it is not completely devoid of genes/cDNAs. The presence of high proportions of middle and low-copy repetitive sequences in this region may have evolutionary significance in the genome organization and function of this region. Since 21q11–21 is heavily methylated, the expression of genes in this region may be regulated by a delicate balance of methylation and demethylation, and the presence of an additional copy of chromosome 21 may seriously disturb this balance and cause specific Down syndrome anomalies including mental retardation.
Keywords: microdissection, Down syndrome, methylation, cDNA, repetitive sequences
The proximal half of the long arm of human chromosome 21, 21q11–21, is a unique chromosomal region in the human genome. This region contains approximately 20 megabases (Mb) of DNA and includes a large dark band of 21q21, plus a small light band of 21q11.2 and the centromeric region 21q11.1. The 21q11–21 region is known for: (i) late replication, (ii) DNase insensitivity, (iii) low activity in recombination and transcription, (iv) fewer natural and induced breakpoints, (v) low GC content and few CpG islands (1–5), (vi) association with Down syndrome (DS) pathologies, including mental retardation (6), and Usher syndrome type I (congenital hearing loss) (7).
Despite intensive efforts, the 21q11–21 region has been refractory to detailed molecular analysis, partly due to the difficulty in isolating adequate molecular probes by conventional molecular cloning, and also due to the lack of effective gene identification and cDNA isolation procedures. For example, plasmid and phage clones isolated from chromosome 21 were mapped largely to the distal half of the long arm of chromosome 21, including 21q22.1-q22.3 bands (8–10). Also, yeast artificial chromosome clones with large inserts from 21q11–21 were found to display large gaps, indicating the presence of unstable sequences (11). In addition, an exhaustive search for cDNAs by hybridization selection procedure using 15 yeast artificial chromosome clones representing a significant portion of 21q11–21 failed to find any expressed genes or cDNAs (12). Similar frustrating experiences were also reported by others (13–15). Thus, this region has been thought to be extremely gene poor or genetically “dead.”
In this study, we used a different approach to isolating useful genomic probes and potential genes from the 21q11–21 region. Employing our previously developed and improved microdissection and MboI linker–adaptor microcloning techniques (16–19), we constructed four microdissection libraries specifically for this region and isolated large numbers of unique sequence microclones. We then sequenced the microclones and searched in databases to identify homologies with known genes and cDNAs. We also used unique sequence microclones in direct screening of cDNA libraries to isolate cDNAs for this region. Here we report that after analyzing more than 2,500 microclones from these libraries we surprisingly found that this region is enriched with middle and low-copy repetitive sequences (>40%), which may be a hindrance to the isolation of regions corresponding to coding sequences by methods like cDNA hybridization selection. By sequencing and homology analysis, we identified a significant number of genes and cDNA correspondences from 21q11–21. This is contrary to the general belief that this region is extremely deficient in genes. We also found that this region is heavily methylated. Thus, the expression of the genes in this region may be regulated by a delicate balance of methylation and demethylation, and the presence of an additional copy of chromosome 21 may seriously disturb this balance and cause specific DS anomalies including mental retardation.
MATERIALS AND METHODS
Chromosome Microdissection and MboI Linker–Adaptor Microcloning.
Giemsa–trypsin banded metaphase chromosomes from human lymphoblast cell line GM03714 (46, XX, Cell Repository, Camden, NJ) were used in microdissection of the 21cen-q21 region using our standard techniques (16–19). Twenty-chromosome fragments were microdissected and pooled in a microdrop of proteinase K solution of 5 nl, which was overlayered with paraffin oil to prevent evaporation. The pooled chromosome fragments were digested with proteinase K, extracted three times with phenol, cleaved with either MboI or Sau3A, ligated to an MboI linker–adaptor. All these steps were carried out in nanoliter microdrops visualized through a microscope and aided by a de Fonbrune micromanipulator, as described (16–19). The ligated sequences were transferred to a 200-μl microtube and amplified for 15 cycles by the polymerase chain reaction (PCR) using the 20-mer sequence of the MboI linker–adaptor molecule as primer (16). After PCR, samples were taken from the resulting PCR products and amplified again for 20 additional cycles to make a final DNA library.
Construction of Microdissection Libraries and Isolation of Unique Sequence Microclones.
Four microdissection libraries were constructed using (i) either MboI or Sau3A as cleavage enzymes for microcloning and (ii) either DH5α or DH10B (BRL) as host cells for transformation. After transformation, white colonies were isolated and hybridized to 32P-labeled total human DNA to identify unique or repetitive sequence inserts in microclones. The conditions for colony hybridization were set to detect microclones with highly repetitive sequences. Colonies with no or very weak hybridization signals, presumably unique or low copy number sequences, were isolated for further analysis.
Analysis of Unique Sequence Microclones.
Unique sequence microclones were analyzed for insert size, human genomic HindIII hybridizing fragment size, human origin, and chromosome 21 specificity. In these analyses, Southern hybridization was used in which labeled microclones were hybridized to DNAs from (i) human (GM03714), (ii) human/mouse cell hybrid WAV17 containing a single human chromosome 21 or human/Chinese hamster cell hybrid R451 (strain R451–29c-5) containing a single human chromosome 21 (provided by Carol Jones, Eleanor Roosevelt Institute), and (iii) mouse A9 or Chinese hamster CHO-K1 cells, as described (16–19).
Refined Regional Mapping of Microclones to Subregions of 21q11–21.
Refined regional mapping of microclones to subregions of 21q11–21 was carried out using the following human/Chinese hamster cell hybrids kindly supplied by David Patterson (Eleanor Roosevelt Institute), as described (20): (i) JC6, (ii) ACEM, (iii) 6918, (iv) 6;21 (R50–3), (v) 4;21 (GA9–3), (vi) 3;21 (9528C-1), and (vii) 1;21 (1881C-13b). Southern hybridization was used for all these regional mapping studies (10).
Methylation Analysis in 21q11–21.
Methylation sensitive enzyme HpaII and its methylation insensitive isoschizomer MspI were used in the methylation analysis of the genomic regions flanked by these enzyme sites. HpaII can recognize and cleave the CCpGG sequence in which the C residue preceding G is not methylated, but cannot cleave the sequence if the C in CpG is methylated. MspI can cleave the CCpGG sequence regardless of the methylation condition in the C. In these studies, individual unique sequence microclones were labeled and hybridized to total human DNA cleaved with either HpaII or MspI to detect differences in genomic fragment sizes resulting from methylation at the HapII site.
DNA Sequencing.
Applied Biosystems automated sequencer 373 was used for sequencing of microclones. Plasmids or amplified inserts from microclones were purified for sequencing. The Applied Biosystems PRISM cycle sequencing reaction kits with ampliTaq DNA polymerase FS (Perkin–Elmer) were used for sequencing reactions.
Identification of Human Genes and cDNAs by Sequence Homology Search in Databases.
Sequences from microclones were searched for homology with known genes and cDNAs using the blast searching strategy for DNA and protein searches (21). The cutoff P value was <1.0e-16 for both DNA and protein sequences. Microclones with homologies to gene sequences in DNA (blastn) only, but not in protein (blastx), were not included in Table 1. Similarly, microclones homologous to genomic sequences like cosmids, sequence-tagged sites, tandem repeats, etc., were not included.
Table 1.
Homology analysis of microclones by sequencing and database search
| Microclone (bp) | Homology hit | blastn (DNA) | blastx (Protein) | Subregion mapping |
|---|---|---|---|---|
| Homology with known human genes | ||||
| SB-487 (316) | Human extracellular signal-regulated kinase 3 (ERK3) | 4.8e-93 | 4.5e-49 | VI |
| SA-103 (279) | Human farnesyl pyrophosphate synthetase* | 1.8e-79 | 4.3e-40 | V |
| MB-4 (474) | Human α-tubulin† | 2.7e-53 | 1.4e-30 | VI |
| MB-42 (189) | Human α-tubulin‡ | 3.5e-33 | 3.1e-16 | VI |
| SB-97 (187) | Human microsomal stress 70 protein ATPase core (STCH)§ | 1.6e-69 | 1.1e-26 | II |
| SB-494 (326) | Human zinc finger/leucine zipper | 7.8e-57 | 1.5e-24 | |
| SB-66 (213) | Human cytokeratin 18 | 1.1e-50 | 2.2e-17 | V |
| Homology with human cDNAs¶ | ||||
| SA-292 (269) | 112113 | 1.3e-68 | — | IV |
| MB-8 (195) | c-38h09 | 2.1e-43 | — | II |
| MA-45 (187) | JJ5383 | 5.0e-30 | — | one of two copies on chromosome 21 mapped to III |
Also hit in Saccharomyces Genome Database SGB, P = 8.2e-11.
Also hit in Saccharomyces Genome Database SGB, P = 1.0e-24.
Also hit in Saccharomyces Genome Database SGB, P = 1.8e-12.
Known to be expressed and mapped to chromosome 21.
Searched in expressed sequence tags database.
Direct Screening of cDNA Libraries Using Unique Sequence Microclones from 21q11–21.
The insert from each unique sequence microclone was individually amplified, pooled in groups of 10 microclones per group, labeled, and used as probes to screen a fetal brain cDNA library (CLONTECH). About 1–2 × 106 plaque-forming units of cDNA were screened with each group of probes. After hybridization, positive plaques were isolated and purified by secondary and tertiary screening, as previously described (22).
Northern Blot Analysis of Microclones.
Multiple tissue Northern blots from CLONTECH (both fetal and adult tissues) were used to hybridize to cDNAs isolated by direct screening according to manufacturer’s instructions.
RESULTS
Construction and Characterization of Microdissection Libraries for 21q11–21.
Four microdissection libraries were constructed using 20 dissected chromosome fragments from 21q11–21 for each library. According to the combinations of the enzymes and bacterial hosts used, these libraries were designated as: (i) MA library (MboI/DH5α), (ii) MB library (MboI/DH10B), (iii) SA library (Sau3A/DH5α), and (iv) SB library (Sau3A/DH10B). Colony hybridization of over 2,500 microclones from the 4 libraries showed that 40–50% (mean 46%) of the clones contained highly repetitive sequences. The insert sizes ranged between 50–600 bp (mean 300 bp). More than 80% of the microclones were shown to derive from chromosome 21 by Southern hybridization.
Southern Hybridization Analysis of Unique and Low-Copy Sequence Microclones.
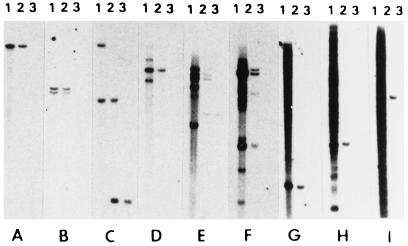
After colony hybridization, which removed about 46% of the highly repetitive sequence microclones, we isolated more than 1,000 microclones from the remaining 54% of the microclones that exhibited no or light hybridization to total human DNA, presumably containing unique or low-copy sequences. In Southern hybridization analysis, we found that these microclones exhibited the following hybridization patterns, as shown in Fig. 1: (i) 12% of the microclones contained unique sequences that displayed unique bands in both human and chromosome 21 (usually 1 band was found but occasionally 2 bands were present, which could result from the presence of an HindIII site within the insert) (Fig. 1 A and B); (ii) 18% contained low-copy sequences showing multiple bands (2 or 3 to 20) or light smear in total human DNA and unique or multiple bands in chromosome 21 (Fig. 1 C–F); (iii) 24% contained middle repetitive sequences that escaped detection in our colony hybridization procedure designed to identify highly repetitive sequences; this class of microclones exhibited relatively dark smear in human DNA but unique or multiple bands or smear in chromosome 21 (Fig. 1 G–I).
Figure 1.
Southern blot hybridization of microclones to total genomic DNAs from (1) human (GM03714), (2) human/Chinese hamster cell hybrid with single human chromosome 21, or (3) Chinese hamster cell line CHO-K1. The following microclones were tested: (A) MB-14, (B) SB-31, (C) MB-69, (D) MA-6, (E) MA-85, (F) MA-235, (G) SB-104, (H) SB-94, and (I) SB-24.
Evolutionary Conservation in Rodent Species.
Microclones were analyzed for homology with rodent species. Among 400 microclones analyzed, 76 (19%) showed cross-hybridization with rodent DNA (either Chinese hamster or mouse or both) (Fig. 1 C and E). Because we used our standard stringent hybridization conditions in these experiments, some microclones with weaker homologies would not be detected.
Refined Regional Mapping of Unique Sequence Microclones Using Cell Hybrids and Southern Hybridization.
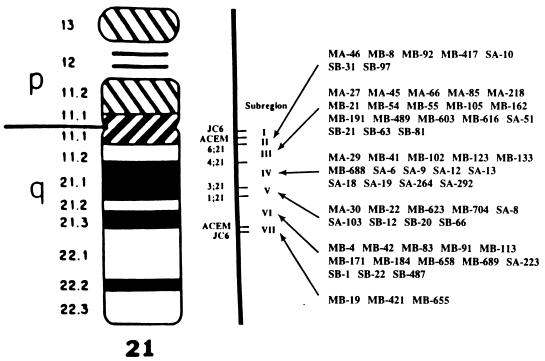
Microclones were hybridized to blots containing DNAs from various hybrids containing parts of chromosome 21. Fig. 2 summarizes the regional assignments of 64 unique sequence microclones derived from the microdissection libraries. It can be seen that the number of microclones assigned to 21q11–21 has been greatly enriched, as compared with those previously described (8–10).
Figure 2.
Refined regional mapping of 64 microclones to subregions within 21q11–21 by Southern hybridization to a chromosome 21 regional mapping panel comprising eight cell hybrids that divide 21q11–21 into seven subregions.
Methylation Analysis.
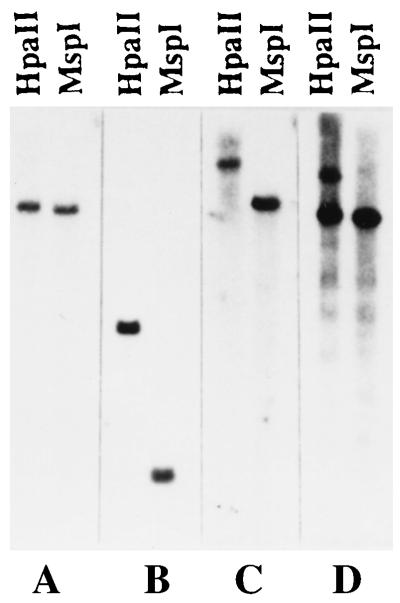
Fig. 3 shows the human genomic fragment sizes cleaved by either HpaII or MspI using unique sequence probes from 21q11–21. Only microclones that showed single unique bands on human DNA were used. Among 43 microclones analyzed, 39 clones (90.7%) showed methylation patterns in the probed region and only 4 clones (9.3%) exhibited no apparent methylation patterns.
Figure 3.
Methylation analysis of microclones hybridized to total human DNA digested completely with either HpaII or MspI. The following microclones were tested: (A) SB-18 (6.0 kb for both HpaII and MspI), (B) SB-1 (1.6 kb for HpaII and 0.6 kb for MspI), (C) SA-11 (14.0 kb for HpaII and 4.6 kb for MspI), (D) SB-31 (8.0 and 4.6 kb for HpaII and 4.6 kb for MspI).
Identification of Human Genes/cDNAs in 21q11–21 by Sequence Homology Search.
Complete sequencing was done for microclones with inserts of 400 bp or less, and partial sequencing from 5′ end was done for longer inserts. About 400 microclones from 21q11–21 were sequenced and searched in databases; 10 were found with high homologies to known genes or cDNAs (Table 1). Only microclones with high homologies (P = <1.0e-16) to both DNA and protein sequences were included. Microclones that have homologies with genes in DNA only, or protein only, were not included. For the cDNA match, only the DNA sequence was used.
Six microclones have high homologies with five known genes at both DNA and protein levels (Table 1). Interestingly, microclone SB-97 (187 bp insert) matches well (186/187 nt or 99% identity; P = 3.1e-71) with the human STCH gene, which encodes the ATPase core of a microsomal stress 70 protein. This gene is a member of the stress 70 chaperon family involved in the ATP-dependent processing of cytosolic and secretory proteins. The STCH gene was previously mapped to 21q11.2 (23) and shown to be expressed in many cell types. In this study, we also demonstrated its expression in multiple tissues with two messages (4.2 and 2.1 kb) detected by SB-97 (the same as the two bands detected by cDNA clone CSA in Fig. 4E). In addition, our regional mapping not only confirmed the previously reported location of STCH between breakpoints 2Fur1 and 6;21 in 21q11.1 (23), but further refined the location to lie between breakpoints ACEM and 6;21 (Fig. 2). SB-97 yielded a single 14-kb HindIII band in both human and chromosome 21 DNA, with several extra bands in human DNA only.
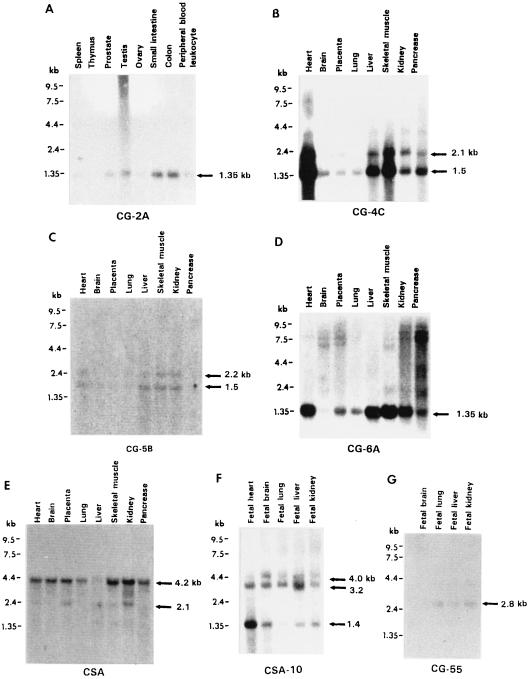
Figure 4.
Northern blot analysis of seven cDNA clones hybridized to multiple tissue Northern blots (CLONTECH) to demonstrate the expression and mRNA size of the cDNAs.
Microclone SA-103 corresponds (with 165/187 nt or 88% homology) to the gene for farnesyl pyrophosphate synthetase (FPS), an enzyme involved in cholesterol biosynthesis. It yielded a 9.6-kb band in both human DNA and chromosome 21, with four extra bands in human DNA only, indicating that several gene family members on other chromosomes as previously described (24, 25).
Two microclones, MB-4 and MB-42, correspond to the gene for α-tubulin, in different parts of the gene. About 15–20 dispersed human genes code for α-tubulin. Both MB-4 and MB-42 yielded a single 20–30-kb band on chromosome 21, with 7–8 extra bands in human DNA only, suggesting that the 2 microclones code for the same α-tubulin gene on chromosome 21. Northern blot analysis of MB-4 detected a 1.4-kb band in many tissues tested (data not shown).
Microclone SB-66 corresponds (with 167/191 nt or 88% homology) to the gene for human cytokeratin 18, a class of intermediate filament subunit proteins. There are at least 30 distinct cytokeratins typically expressed in epithelial cells. SB-66 yielded a single band in chromosome 21 and multiple bands in human DNA.
Microclone SB-487 corresponds (with 181/199 nt or 90% homology) to the gene for human extracellular signal-regulated protein kinase 3 (ERK3), a member of the gene family involved in converting tyrosine phosphorylation into the serine/threonine phosphorylation cascades (26). These genes may be activated in response to a wide variety of extracellular stimuli, and individual family members may exert unique actions in different cell types or during different developmental stages. SB-487 yielded a single dark band at 6.5 kb in both human DNA and chromosome 21, plus two light bands in human DNA only, and detected a 4.3-kb mRNA band in fetal heart, brain, lung, liver, and kidney (data not shown).
Microclone SB-494 corresponds (with 187/227 nt or 82% homology) to the gene for human zinc finger/leucine zipper protein AF10, a novel class of transcription factors (27). AF10 encodes a protein with an N-terminal zinc finger region and a C-terminal leucine zipper. This gene is frequently involved in the t(10;11)(p12;q23) translocation of acute myeloid leukemias (27). SB-494 yielded six dark bands in human DNA and a single light band in chromosome 21. Each of the dark bands in human DNA could represent a cluster of several copies of the gene. However, chromosome 21 appears to have only one copy.
Three microclones have identical or near-identical homologies with previously reported cDNAs in the database. Microclone MB-8 yielded a single 5.4-kb band in human DNA and chromosome 21. It is identical to cDNA C-38h09 (P = 2.1e-43), which was previously isolated by Genethon from the normalized infant brain cDNA library of B. Soares (Columbia University) but not mapped. The 195-bp insert in MB-8 has 100% homology with the 345-bp cDNA. Thus, this cDNA can now be assigned to chromosome 21. Using the cell hybrid mapping panel, we further localized this cDNA to Subregion II, within ACEM and 6;21 breakpoints (Fig. 2). MB-8 showed positive expression in fetal liver (8.5 kb), but not in fetal brain, lung, and kidney (data not shown). Microclone MA-45 yielded two bands (3.8 and 2.9 kb) in both human and chromosome 21 DNA, with several extra bands in human DNA only. We mapped the 3.8-kb band to Subregion III. This microclone is almost identical (86/88 nt or 97% homology) to cDNA JJ5383 (173 bp) isolated from a fetal heart cDNA library (University of Toronto). Microclone SA-292 yielded a single 6.4-kb band in human and chromosome 21 DNA. No other bands were found in human DNA. It is nearly identical (94–100% homology for different parts of the sequence) to cDNA 112113 (386 bp) isolated from a fetal liver spleen cDNA library of B. Soares, and sequenced by the Washington University–Merck EST Project (accession no. T91946). Northern blot analysis showed that MA-45 detected a 1.35-kb band in all tissues tested (the same as the band detected by cDNA clone CG-6A in Fig. 4D).
Overall, among the nine genes/cDNAs identified by homology analysis (Table 1), at least four genes/cDNAs are certain to be expressed and encoded by 21q11–21: SB-97/STCH, SB-487/ERK3, MB-8/c-38h09, and MA-45/JJ5383. Two other microclones possibly encode expressed genes/cDNAs on 21q11–21: MB-4/α-tubulin and SA-292/112113. The remaining three microclones, SA-103/FPS, SB-494/zinc finger, and SB-66/cytokeratin 18, failed to show detectable expression so far and require further verification.
Isolation of cDNA by Direct Screening of a Fetal Brain cDNA Library Using Unique Sequence Microclones from 21q11–21.
After screening a cDNA library with 120 unique sequence microclones, 12 positive plaques were isolated and purified. Seven of these cDNAs analyzed thus far exhibited unique sequence patterns on chromosome 21. They were further mapped to the following refined regions within 21q11–21: CG-4C (insert 1.0 kb) to Subregion III, CG-5B (2.3 kb) to Subregion III, CG-6A (1.1 kb) to Subregion III, CG-55 (1.5 kb) to Subregion III, CSA (3.0 kb) to Subregion II, CSA-10 (1.8 kb) to Subregion II, and CG-2A (2.2 kb) to 21q11–21. Two of these cDNAs were also identified by the following microclones: CG-6A identified by microclone MA-45/JJ5383 and CSA identified by SB-97/STCH. In addition, CSA-10 was identified by microclone SA-10 and CG-55 identified by MB-55. A database search of the partial sequences of four of these cDNAs (CG-2A, CG-5B, CG-6A, and CG-55) revealed no homology with known genes/cDNAs, and is thus likely novel.
Northern Blot Analysis of cDNAs for Tissue Expression and mRNA Size Determination.
The above seven cDNAs were analyzed by Northern hybridization and the results are shown in Fig. 4. It can be seen that these cDNAs are expressed in various tissues in different amounts, from which the mRNA sizes were determined.
DISCUSSION
Table 2 summarizes the genomic analysis of the 21q11–21 region using more than 2,500 microclones from the libraries specifically constructed for this region. The 21q11–21 region has been generally regarded as gene poor since it contains relatively lower GC content and fewer CpG islands (2–5), and only five genes and very few cDNAs have so far been identified for this region of approximately 20 Mb (12–15, 23, 28–31). In contrast, more than 50 genes and over 200 cDNAs have been identified in the 21q22 region of similar size (ref. 32 and §). Our study employing homology search and direct cDNA screening by using a fraction of the unique sequence microclones from 21q11–21 has uncovered 18 potential genes/cDNAs, of which at least nine are certain to be expressed and encoded by 21q11–21. These nine genes/cDNAs include: 4 (of 9) from homology analysis and 7 (of 12) from cDNA screening; of these 11, 2 derived from homology analysis were the same as the 2 cDNAs isolated from cDNA screening. Thus, while it is true that 21q11–21 may be poor in housekeeping genes as reflected by fewer CpG islands, this region appears not totally deficient in genes, especially gene family members, and the genes may not be all silent.
Table 2.
Summary of the genomic analysis of the 21q11-21 region
| Genomic size of 21q11-21 | ∼20 Mb |
| Microdissection libraries specific for 21q11-21 | MA, MB, SA, SB |
| Independent, individual clones in the libraries | >40,000 (mean insert size 300 bp) |
| Cloned genomic size for 21q11-21 | >12 Mb |
| No. of microclones analyzed | >2,500 by repetitive sequence screening |
| >1,000 by Southern hybridization | |
| >400 (>120 kb DNA) by DNA sequencing | |
| Highly repetitive sequences in 21q11-21 | 46% |
| Moderately repetitive sequences in 21q11-21 | 24% |
| Low-copy sequences in 21q11-21 | 18% |
| Unique sequences specific for 21q11-21 | 12% |
| Methylation in 21q11-21 (HpaII sites) | >90% |
| Conserved microclones (cross-hybridizing to rodents) | 19% (76/400) in unique and low-copy microclones |
| GC content in unique and low-copy microclones | <40% |
Another significant finding in this study is the high degree of methylation in the 21q11–21 region as revealed by the use of unique sequence microclones from 21q11–21, which detected methylation patterns in over 90% of the region. This is in contrast to the general methylation levels of about 60% in mammalian genomes (33). The high degree of methylation in this region may lead to chromatin condensation as shown in 21q11–21 (34), which in turn could explain some of the unique features observed in this region, i.e., late replication, DNase insensitivity, low activity in recombination and in transcription, and fewer natural and induced breakpoints (1). Chromatin condensation in 21q11–21 could also lead to gene sequestration and inactivity (34). More importantly, the genes in this region may be readily rendered inactive by mechanisms like methylation, and also become active by demethylation. This simple and direct regulatory mechanism is particularly useful for the gene family members whose expression appears to depend on special needs or during specific developmental stages. In this study, the genes identified by sequence homology analysis are mostly as gene family members in which methylation may play an important role in regulation. In addition, the other five genes previously assigned to 21q11–21 are also members of gene families including (i) APP in the amyloid precursor protein family (28), (ii) enterokinase (serine protease 7) in the serine protease family (29), (iii) NCAM-21 in the neural cell adhesion molecule family (30), (iv) RAP140 in the transcription factor family (31), and (v) STCH in the stress 70 chaperon family (23). However, whether these genes are regulated by methylation remains to be elucidated.
The 21q11–21 region contains as much as 24% middle repetitive and 18% low-copy sequences. The presence of such high proportions of middle and low-copy repetitive sequences in 21q11–21, together with the high level of methylation in this region, may have evolutionary significance in the genome organization, stability, and function of this region (35, 36). About 12% (out of 18% total) of the low-copy repeats analyzed in this study were homologous to endogenous movable sequences including retroviral transposable elements. Thus, the methylation pattern in this region might be initiated by the invasion of such movable elements that activated a methylation-silencing defense system against parasitic sequences in the human cell (35). On the other hand, the high degree of methylation in 21q11–21 may offer a mechanism to stabilize the structure of this region during evolution by diversifying duplicated sequences through mutation (e.g., from methylated cytosine to thymine), resulting in the reduction of recombination-mediated chromosome rearrangements (36). The complex repetitive sequences in 21q11–21 may also cause ineffective isolation of cDNAs by methods such as cDNA hybridization selection (12–15). Because we used only unique sequence microclones in our direct cDNA screening, it might have overcome this sequence complexity and yielded cDNAs from this region.
Due to the high mutation rate of methylated cytosine to thymine (C to T transition) by deamination (37), it is reasonable to assume that hypermethylation in 21q11–21 may lead to CpG dinucleotide depletion and may result in a region low in GC content and CpG islands. In accordance with this interpretation, extensive studies have shown that 21q11–21 is indeed GC poor and with few CpG islands (1–5). Furthermore, recent studies by hybridization selection using yeast artificial chromosome and cosmid clones spanning a large portion of 21q11–21 uncovered only a few genes or cDNAs (12–15). Thus, this region has been generally viewed as a desert for genes/cDNAs and is perhaps full of meaningless sequences. However, our studies demonstrated that this region, although not gene rich, is not completely devoid of novel genes, particularly gene family members. Moreover, the high level of methylation in this region may play a unique role in the regulation of expression of the genes in this region, particularly during early development (38). Since certain DS abnormalities, including mental retardation, are associated with 21q11–21, the genes residing in this region should be carefully examined. In particular, the presence of three copies of genes in DS individuals may seriously disturb the delicate balance in methylation and demethylation of these genes during critical embryonic development stages and may result in specific DS pathologies.
Acknowledgments
We thank J. Shu for excellent technical assistance. We are also grateful to Drs. David Patterson and Carol Jones for providing cell hybrids and to Dr. Katheleen Gardiner for valuable discussions. This study was supported by National Institutes of Health Grant HD-17749.
ABBREVIATIONS
- DS
Down syndrome
- Mb
megabase
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF583–AF597).
The Sixth International Workshop on Human Chromosome 21, Cold Spring Harbor Laboratory, NY, May 6–8, 1996.
References
- 1.Patterson D, Epstein C J, editors. Molecular Genetics of Chromosome 21 and Down Syndrome. New York: Wiley–Liss; 1990. [Google Scholar]
- 2.Gardiner K, Horisberger M, Kraus J, Tantravahi U, Korenberg J, Rao V, Reddy S, Paterson D. EMBO J. 1990;9:25–34. doi: 10.1002/j.1460-2075.1990.tb08076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardiner K, Aissani B, Bernardi G. EMBO J. 1990;9:1853–1858. doi: 10.1002/j.1460-2075.1990.tb08310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassone F, Cheng S, Gardiner K. Am J Hum Genet. 1992;51:1251–1264. [PMC free article] [PubMed] [Google Scholar]
- 5.Saccone S, De Sario A, Valle G D, Bernardi G. Proc Natl Acad Sci USA. 1992;89:4913–4917. doi: 10.1073/pnas.89.11.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korenberg J R, Chen X-N, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham J M, Jr, Hugdins L, McGillivray B, Miyazaki K, Ogasawara N, Park J P, Pagon R, Pueschel S, Sack G, Say B, Schuffenhauer S, Soukup S, Yamanaka T. Proc Natl Acad Sci USA. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaib H, Kaplan J, Gerber S, Vincent C, Ayadi H, Slim R, Munnich A, Weissenbach J, Petit C. Hum Mol Genet. 1997;6:27–31. doi: 10.1093/hmg/6.1.27. [DOI] [PubMed] [Google Scholar]
- 8.Korenberg J R, Croyle M L, Cox D R. Am J Hum Genet. 1987;41:963–978. [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Erickson P, Patterson D, Jones C, Drabkin H. Genomics. 1991;10:166–172. doi: 10.1016/0888-7543(91)90497-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Hartz J, Xu Y, Gemmill R M, Korenberg J R, Patterson D, Kao F T. Am J Hum Genet. 1992;51:263–272. [PMC free article] [PubMed] [Google Scholar]
- 11.Gardiner K, Graw S, Ichikawa H, Ohki M, Joetham A, Gervy P, Chumakov I, Patterson D. Somat Cell Mol Genet. 1995;21:309–414. doi: 10.1007/BF02310207. [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Wei H, Tassone F, Graw S, Cardiner K, Weissman S M. Genomics. 1995;27:1–8. doi: 10.1006/geno.1995.1001. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J F, Boyartchuk V, Zhu Y. Genomics. 1994;23:75–84. doi: 10.1006/geno.1994.1461. [DOI] [PubMed] [Google Scholar]
- 14.Yaspo M L, Gellen L, Mott R, Korn B, Nizetic D, Poustka A M, Lehrach H. Hum Mol Genet. 1995;4:1291–1304. doi: 10.1093/hmg/4.8.1291. [DOI] [PubMed] [Google Scholar]
- 15.Tassone F, Xu H, Burkin H, Weissman S, Gardiner K. Hum Mol Genet. 1995;4:1509–1518. doi: 10.1093/hmg/4.9.1509. [DOI] [PubMed] [Google Scholar]
- 16.Kao F T, Yu J W. Proc Natl Acad Sci USA. 1991;88:1844–1848. doi: 10.1073/pnas.88.5.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Qi J, Tong S, Kao F T. Hum Genet. 1994;93:557–562. doi: 10.1007/BF00202823. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, Tong S, Whittier A, Kao F T. Somat Cell Mol Genet. 1995;21:335–343. doi: 10.1007/BF02257468. [DOI] [PubMed] [Google Scholar]
- 19.Kao F T. In: Methods of Genome Analysis in Plants. Jauhar P P, editor. Boca Raton, FL: CRC; 1996. pp. 329–343. [Google Scholar]
- 20.Graw S L, Gardiner K, Hall-Johnson K, Hart I, Joetham A, Walton K, Donaldson D, Patterson D. Somat Cell Mol Genet. 1995;21:415–428. doi: 10.1007/BF02310208. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Kao F T, Yu J, Tong S, Qi J, Patanjali S R, Weissman S, Patterson D. Genomics. 1994;23:700–703. doi: 10.1006/geno.1994.1561. [DOI] [PubMed] [Google Scholar]
- 23.Brodsky G, Otterson G A, Parry B B, Hart I, Patterson D, Kaye F J. Genomics. 1995;30:627–628. doi: 10.1006/geno.1995.1291. [DOI] [PubMed] [Google Scholar]
- 24.Sheares B T, White S S, Molowa D T, Chan K, Ding V D, Kroon P A, Bostedor R G, Karkas J D. Biochemistry. 1989;28:8129–8135. doi: 10.1021/bi00446a025. [DOI] [PubMed] [Google Scholar]
- 25.Heinzmann C, Clarke C F, Klisak I, Mohandas T, Sparkes R S, Edwards P A, Lusis A J. Genomics. 1989;5:493–500. doi: 10.1016/0888-7543(89)90015-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhu A X, Zhao Y, Moller D E, Flier J S. Mol Cell Biol. 1994;14:8202–8211. doi: 10.1128/mcb.14.12.8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaplin T, Ayton P, Bernard O A, Saha V, Della V, Hillion J, Gregorini A, Lillington D, Berger R, Young B D. Blood. 1995;85:1435–1441. [PubMed] [Google Scholar]
- 28.Patterson D, Gardiner K, Kao F T, Tanzi R, Watkins P, Gusella J F. Proc Natl Acad Sci USA. 1988;85:8266–8270. doi: 10.1073/pnas.85.21.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitamoto Y, Veile R A, Donis-Keller H, Sadler J E. Biochemistry. 1995;34:4562–4568. doi: 10.1021/bi00014a008. [DOI] [PubMed] [Google Scholar]
- 30.Paoloni-Giacobino A, Chen H M, Rossier C, Antonarakis S E. Am J Hum Genet. 1995;57:A150. doi: 10.1007/s004390100541. (abstr.). [DOI] [PubMed] [Google Scholar]
- 31.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu M, Antonarakis S E, Van Broeckhoven C, Patterson D, Gardiner K, Nizetic D, Creau N, Delabar J M, Korenberg J, Reeves R, Doering J, Chakravati A, Minoshima S, Ritter O, Cuticchia J. Cytogenet Cell Genet. 1955;70:148–182. [Google Scholar]
- 33.Wigler M H. Cell. 1981;24:285–286. doi: 10.1016/0092-8674(81)90317-2. [DOI] [PubMed] [Google Scholar]
- 34.Puck T T, Johnson R. Stem Cells. 1996;14:548–557. doi: 10.1002/stem.140548. [DOI] [PubMed] [Google Scholar]
- 35.Bestor T, Tycko B. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 36.Kricker M C, Drake J W, Radman M. Proc Natl Acad Sci USA. 1992;89:1075–1079. doi: 10.1073/pnas.89.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selker E U. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 38.Razin A, Shemer R. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]