Abstract
Early meiotic nodules (also called recombination nodules) are proteinaceous structures about 100 nm in diameter that are associated with forming synaptonemal complexes (SCs) during early prophase I of meiosis. Although their function is unknown, early nodules may be involved in searching for DNA homology before synaptic initiation. Two potential components of early nodules are Rad51 and Dmc1 proteins. These proteins are important for meiotic recombination in eukaryotes and are homologous to RecA, the major protein that catalyzes homologous pairing and DNA strand exchange in prokaryotes. In addition, Rad51 has been localized by immunofluorescence in abundant foci that may correspond to early nodules in yeast, lily, and mouse. In yeast and lily, Dmc1 and Lim15, the lily homolog of Dmc1, colocalize with Rad51. Here, using electron microscopic immunogold localization to spreads of zygotene and early pachytene SCs from lily, we confirm that RecA-like proteins are components of early nodules. The antibody used was generated to full-length tomato Rad51 protein and binds to both Rad51 and Lim15 in immunoblots of lily primary microsporocyte proteins. The labeled early nodules are heterogeneous in size and are associated with both axial elements and SCs. There are two classes of early nodules, those that are densely labeled with gold and those that are not labeled at all. This result may be due to technical limitations associated with using spread preparations or to differences in the nodules themselves. The presence of Rad51 and/or Lim15 proteins in early nodules supports the hypothesis that early nodules are involved in recombination-related events during meiosis.
During early prophase of the first meiotic division (prophase I), homologous chromosomes come together in pairs, synapse along their lengths by formation of SCs to form bivalents, and recombine (1–3). These events are important both for generating new combinations of genes and for the proper segregation of homologous chromosomes at anaphase I. Meiotic nodules are spherical to ellipsoidal proteinaceous structures approximately 100 nm in diameter that become closely associated with forming and completed SCs during prophase I (3–6). In many eukaryotes, two types of meiotic nodules (early and late) can be distinguished from one another using a combination of the following characteristics: stage of appearance, frequency, shape, size, and staining properties (refs. 5 and 6; L.K.A., unpublished observations). Meiotic nodules are also called recombination nodules (4, 5). We prefer the more generally descriptive term meiotic nodules because the role of early nodules in recombination has not yet been firmly established (6). At leptonema, the stage of prophase I immediately prior to synapsis, numerous early nodules associate with proteinaceous structures called axial elements that form between each pair of sister chromatids. During the process of synapsis at zygonema, early nodules are often observed at sites of convergence between synapsing axial elements of homologous chromosomes as well as in association with completed SCs (7–9). When synapsis is complete at early pachynema, the number of early nodules progressively decreases so that from middle through late pachynema, no early nodules are left. Late nodules appear on the central element of SCs during early pachynema and persist into early diplonema when SCs disintegrate. Normally, every pachytene SC has at least one late nodule, and late nodules are directly correlated with chiasmata and reciprocal recombination events in a number of organisms. This has led several investigators to suggest that late nodules are involved in crossing over (e.g., refs. 4 and 10–14). The function of early nodules is less clear. It has been proposed that some early nodules develop into late nodules (e.g., ref. 15). In addition, it has been suggested that early nodules are involved in synaptic initiation (7–9), homology search, and/or gene conversion (16–17).
One approach to defining the function of meiotic nodules is to identify their protein constituents, particularly with regard to proteins known to be involved in recombination. Genetic and biochemical evidence from Saccharomyces cerevisiae (yeast) indicates that genes of the RAD52 epistasis group (RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, XRS2, and MRE11) are involved in both mitotic and meiotic recombination through DNA double-strand break repair (18–20). Three of the yeast genes (RAD51, RAD55, and RAD57) plus a related meiosis-specific yeast gene (DMC1) code for proteins that are similar to RecA, a protein important for prokaryotic recombination (21–23). Rad51 is the most intensively studied of the RecA-like proteins in eukaryotes. Like RecA, yeast Rad51 protein binds to DNA in an ATP-dependent manner to form nucleoprotein filaments that are necessary for strand exchange between nicked homologous DNA molecules (24–27). Homologs to the yeast RAD51 gene have been identified in several different eukaryotes (e.g., human, mouse, and tomato), and a homolog to the yeast DMC1 gene called LIM15 has been identified in lily (21, 22, 28). The amino acid similarities of the predicted gene products indicate conservation of function (21, 22, 28). Functional conservation of Rad51 is further supported by the partial complementation of certain rad51 mutations in yeast by the mouse RAD51 gene (29). Because yeast rad51 and dmc1 mutants accumulate DNA double-strand breaks during meiosis, Rad51 and Dmc1 proteins are thought to operate after the formation of these breaks, presumably in searching for homology and strand transfer (20). Recent analysis of yeast zip1 rad51 and zip1 dmc1 double mutants indicates that both Rad51 and Dmc1 proteins are important for promoting chromosome synapsis during meiosis (30). Given the biochemical, cytological, and genetic evidence regarding Rad51 and Dmc1 and the proposed function of early meiotic nodules, it is reasonable to hypothesize that these RecA-like proteins are present in early nodules (30–33). Indeed, by using immunofluorescent labeling, Rad51 proteins have been localized in numerous foci along early prophase I chromosomes of yeast, lily, and mouse (31–33). In meiotic prophase nuclei of yeast and lily, Dmc1 and Lim15 both colocalize with Rad51 in foci (31–32). Although it seems likely that the foci correspond to early meiotic nodules (30–33), these immunolocalization studies are not definitive because nodules cannot be resolved directly by light microscopy. Here we present evidence using electron microscopic immunogold localization that these RecA-like proteins are components of early nodules in a higher plant, Lilium longiflorum (lily).
MATERIALS AND METHODS
Lilium longiflorum (Easter lily) and Lycopersicon esculentum (tomato) plants were grown to flowering at 15–20°C in a greenhouse. Because lilies are grown from bulbs that are reproduced clonally, there is little variation between different lily plants. Meiotic material from several different plants was used to prepare squashes and spreads for antibody labeling. Anthers from tomato were used to prepare a cDNA library from which the RAD51 gene was cloned using a portion of the S. cerevisiae DMC1 gene as a probe (28).
Preparation of Affinity-Purified Antibodies to Rad51 Protein.
The complete tomato RAD51 gene was expressed in E. coli, and the full-length fusion protein was purified using a Ni-NTA column (Qiagen, Chatsworth, CA). Polyclonal antibodies were generated in a rabbit by repeated injections with purified Rad51 fusion protein. The antibodies were affinity-purified from immunoblots of the fusion protein or from an affinity column of the Rad51 fusion protein (34).
One- and Two-Dimensional Immunoblots.
Total protein was extracted from isolated lily primary microsporocytes at zygonema and pachynema and from whole tomato anthers containing primary microsporocytes at zygonema and pachynema. For all protein electrophoresis, approximately equal amounts of protein (≥20 μg of total protein) were loaded into each lane or isoelectric focusing gel. For one-dimensional separation, proteins were separated in a 10% SDS polyacrylamide gel (35). A portion of the gel was stained with Coomassie blue, and the remainder of the gel was electroblotted onto nitrocellulose (36). Two-dimensional separation of proteins was performed according to O’Farrell (37), as described by Lammers et al. (38), and then the proteins were electroblotted onto nitrocellulose as described above. The blots were blocked with 5% nonfat dry milk with 5% normal goat serum in TBST (10 mM Tris⋅HCl, pH 8/150 mM NaCl/0.1% Tween-20) for 1 hr, washed in TBST, incubated in affinity-purified polyclonal antibodies to Rad51 diluted 1:100 in blocking solution or polyclonal antibodies to Lim15 diluted 1:4,000 in blocking solution, washed, incubated in goat anti-rabbit antibody conjugated to alkaline phosphatase, and stained using the 5-bromo-4-chloro-3-indoyl phosphate p-toluidine salt/nitroblue tetrazolium procedure (34).
Immunofluorescent and Immunogold Labeling.
Squashes of lily microsporocytes at different stages of meiosis were prepared as described by Terasawa et al. (32). Squashes were incubated in 0.1 M ammonium chloride in PBS for 5 min, washed in PBS for 5 min, blocked with 1% BSA in PBS for 30 min at room temperature, and incubated in affinity-purified antibodies to Rad51 protein diluted 1:10 in blocking solution overnight at 4°C. Squashes were washed with 0.1% acetylated BSA (Aurion, The Netherlands) in PBS and incubated for 1 hr at room temperature in 1:100 dilution of FITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch). Nuclei were counter-stained with 0.2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min. Slides were mounted in Vectashield (Vector Laboratories) and examined with an Olympus AX-70 (New Hyde Park, NY) epifluorescence microscope. Nuclei were photographed at ASA 1600 using T-MAX 400 black and white film.
For immunogold labeling, primary microsporocyte nuclei from several lily plants were used to prepare SC spreads by agar filtration as described by Anderson et al. (39). Some spreads were digested with DNase I (1 μg/ml), and then all spreads were fixed with 4% paraformaldehyde in 50 mM potassium phosphate buffer, pH 7.5. SC spreads on plastic films were transferred to grids and incubated with antibodies as described earlier, except that the secondary antibody incubation was at room temperature for 2 hr with 1:20 dilution of 5 nm colloidal gold conjugated to goat anti-rabbit IgG (Goldmark, Phillipsburg, NJ). Labeled preparations were stained 5 min each with aqueous 2% uranyl acetate and Reynold’s lead citrate and examined in an electron microscope.
Although the antibody was raised to tomato Rad51 protein, we were unable to prepare tomato SCs that were suitable for immunogold labeling due to technical difficulties. However, squashes of tomato microsporocyte nuclei at early stages of prophase I labeled with affinity-purified antibody to Rad51 protein resulted in numerous Rad51 foci similar to our observations for lily.
RESULTS
Characterization of Affinity-Purified Antibodies to Rad51 Protein.
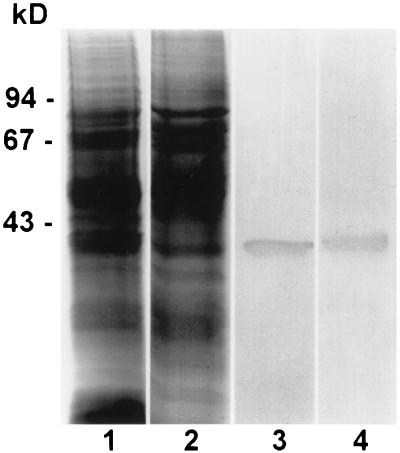
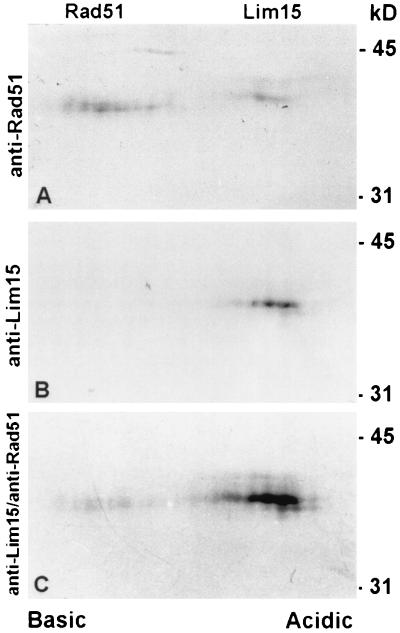
Affinity-purified antibodies to tomato Rad51 protein bind to one band on immunoblots of total protein extracts from lily primary microsporocytes and tomato anthers separated by SDS/PAGE in one dimension (Fig. 1). Comparable immunoblots of lily and tomato meiotic proteins incubated with preimmune serum resulted in no staining of the blot in the region of interest. The stained protein band has an electrophoretic mobility corresponding to a molecular mass of 40 kDa, which is close to the 37-kDa size predicted from the sequence of the tomato RAD51 gene (28). Because the antibodies were elicited to full-length Rad51 protein, we thought it was possible that the antibodies would bind to other RecA-like proteins such as Lim15, the lily homolog of Dmc1. However, immunoblots incubated with antibodies specific to the nonconserved N terminus of Lim15 protein (32) or with a combination of antibodies to Rad51 and Lim15 still stained only one protein band at 40 kDa. Because the predicted molecular masses of Rad51 and Lim15 (38 kDa) proteins differ by only 1 kDa and may not be resolved using a one-dimensional gel, we tested the antibodies on immunoblots of two-dimensional gel electropherograms of total protein extracts from lily primary microsporocytes (Fig. 2). Two of the two-dimensional separations were performed in parallel. For one immunoblot, affinity-purified antibodies to Rad51 bind to two proteins, a more basic protein of approximately 40 kDa and a more acidic protein of approximately 41 kDa (Fig. 2A). The two proteins are separated by less than 1 pH unit. The other blot was immunostained with antibodies to the nonconserved N terminus of Lim15 protein (Fig. 2B), then the blot was subsequently immunostained with affinity-purified antibodies to Rad51 (Fig. 2C). Based on a comparison of the molecular masses and pIs predicted for tomato Rad51 protein (37 kDa; pI = 5.9) and lily Lim15 (38 kDa; pI = 5.5), the protein on the left of the blot corresponds to Rad51 and the protein on the right of the blot corresponds to Lim15. Therefore, our affinity-purified antibodies to full-length tomato Rad51 protein bind to two RecA-like proteins from lily primary microsporocytes, Rad51 and Lim15. For simplicity, we will call this antibody anti-Rad51 with the understanding that structures labeled with this antibody could contain Rad51, Lim15, or a combination of both proteins.
Figure 1.
Extracts of total protein from lily primary microsporocytes at zygonema-pachynema (lanes 1 and 3) and tomato anthers containing primary microsporocytes at zygonema-pachynema (lanes 2 and 4) separated in one dimension and stained with Coomassie blue (lanes 1 and 2) or immunostained with affinity-purified polyclonal antibodies to full-length tomato Rad51 protein (lanes 3 and 4). In each immunoblot, the antibodies bind to a single protein band of approximately 40 kDa. From DNA sequence analysis, the predicted mass of tomato Rad51 protein is about 37 kDa (28). Molecular size standards are indicated on the left in kilodaltons.
Figure 2.
Total protein from lily primary microsporocytes separated in two dimensions, blotted, and immunostained with affinity-purified antibodies to Rad51 (A) or antibodies to the nonconserved N terminus of Lim15 (B). (C) The blot from B was subsequently immunostained with affinity-purified antibodies to Rad51. In A, affinity-purified antibodies to Rad51 protein bind to two proteins that migrate at 40 kDa and 41 kDa, which are separated by less than 1 pH unit. In the second blot (B and C), antibodies to Lim15 bind to the more acidic protein and subsequent staining of the same blot with antibodies to Rad51 not only intensifies the stain of the Lim15 protein, but also reveals the more basic Rad51 protein. Based on these results and the predicted molecular masses and pIs for Rad51 (37 kDa; pI = 5.9) and Lim15 (38 kDa; pI = 5.5), the protein on the left corresponds to Rad51 and the protein on the right corresponds to Lim15. Molecular size standards are indicated on the right in kilodaltons.
Immunofluorescent Labeling with Affinity-Purified Antibodies to Rad51.
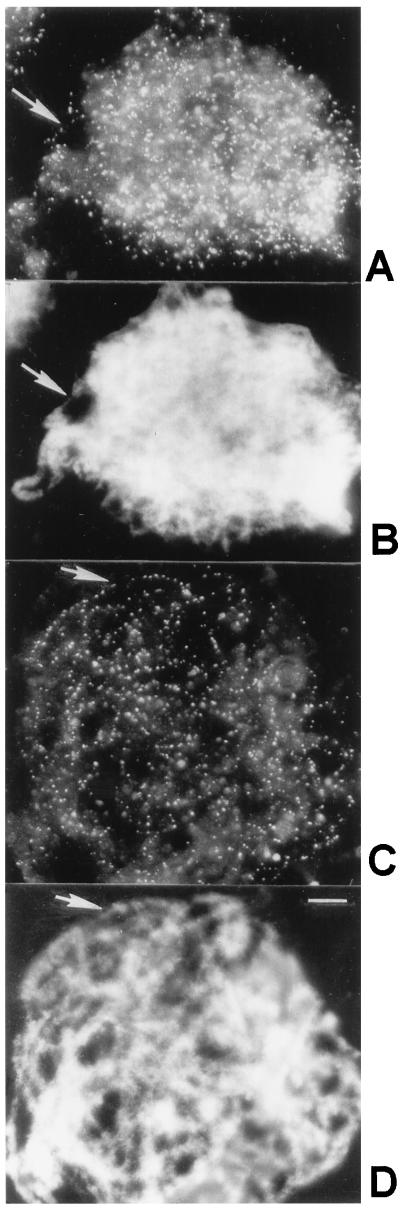
Immunofluorescent labeling of squashed lily primary microsporocytes at the prophase I stages of leptonema and zygonema revealed numerous Rad51 foci as demonstrated previously by Terasawa et al. (32) for lily primary microsporocytes (Fig. 3 A and B). In addition, we observed many Rad51 foci along chromosomal axes at early pachynema (Fig. 3 C and D). Negative controls in which squashed nuclei were incubated in the absence of primary antibody and with preimmune serum resulted in no labeling of microsporocyte nuclei (not shown). Our results are similar to those described earlier for yeast and mouse regarding the immunofluorescent localization of Rad51 protein (31, 33).
Figure 3.
Squashed lily primary microsporocytes at zygonema (A and B) and early pachynema (C and D). (A and C) Immunofluorescent images using affinity-purified antibodies to Rad51 protein and secondary antibodies conjugated to fluorescein isothiocyanate. (B and D) DAPI images of the corresponding nuclei. Arrows in A and B show that individual foci follow unsynapsed chromosomes, and arrows in C and D show that foci also follow synapsed chromosomes. The squashes are not completely flat, and many Rad51 foci are not in the plane of focus. (Bar = 10 μm.)
Immunogold Labeling with Affinity-Purified Antibodies to Rad51.
Although lily has excellent SC and nodule morphology by electron microscopy, the genome is so large (1C = 35 pg) that chromatin can interfere with the penetration of relatively large gold-conjugated antibodies to underlying SCs and nodules in spread preparations (39). Previous experiments labeling lateral elements from lily with 10-nm colloidal gold particles demonstrated that DNase digestion was an effective method to improve the efficiency of labeling (39). To test the effects of DNase digestion on immunolabeling with antibodies to Rad51, we prepared SC spreads in the same way as for electron microscopy but incubated the spreads with FITC-conjugated secondary antibody. After DNase digestion, nuclei did not stain with DAPI, indicating that practically all DNA had been removed. We found that the number, size, and fluorescent intensity of Rad51 foci in spreads digested with DNase were similar to those not digested with DNase (figure not shown). In addition, there was no noticeable ultrastructural difference between nodules in DNase-treated or untreated preparations (see also ref. 39). Comparable experiments with gold-conjugated secondary antibody viewed by electron microscopy showed that DNase digestion greatly increased the numbers of labeled nodules, presumably by allowing better access of gold-conjugated antibodies to the primary antibodies. Therefore, all immunogold observations were made from lily SC spreads that had been treated with DNase I prior to fixation and immunolabeling.
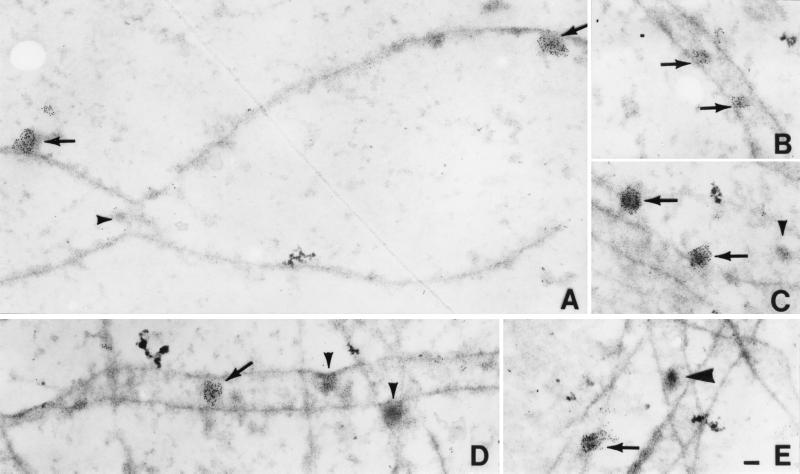
Numerous early nodules were observed by electron microscopy in spreads of lily nuclei at mid- to late zygonema and early pachynema (Fig. 4). Many, but not all, of the nodules were labeled with affinity-purified antibodies to Rad51 protein. The labeled early nodules are heterogeneous in size, ranging from 35 × 70 nm to 140 × 200 nm, and are associated with both axial elements and SCs. At one clear axial element convergence, the nodule at the convergence is not labeled whereas two other nodules associated with an axial element are labeled (Fig. 4A). However, it is unclear whether this is typical because we observed only six axial element convergences and the nodules at two of these were labeled. The density of gold particles on labeled nodules indicates that the nodules contain many Rad51 and/or Lim15 protein molecules. The average number of gold particles on early nodules from zygonema was 51 (SD = 27, n = 36) and on early nodules from pachynema was 72 (SD = 51, n = 26). Unfortunately, little is known about the quantitative relationship between the absolute number of protein molecules and gold grains using immunogold labeling (40), so we cannot estimate the absolute number of Rad51 protein molecules per nodule from the labeling intensity. However, it seems unlikely that there is any amplification of the signal by the indirect immunogold labeling method (40).
Figure 4.
Immunogold labeling of spread SCs from lily using affinity-purified antibodies to Rad51 protein and secondary antibodies conjugated to 5-nm gold particles. (A) Portion of two converging axial elements from a mid- to late zygotene nucleus. Two early nodules associated with one axial element are labeled (arrows), but a small nodule, present at the site of convergence, is not labeled (arrowhead). (B) Portion of an SC from a zygotene nucleus with two small labeled early nodules (arrows). (C) An axial element and an SC from a zygotene nucleus with two early nodules that are abundantly labeled (arrows) and a smaller nodule that is not labeled (arrowhead). (D) Portion of a zygotene nucleus with one SC that extends horizontally across the figure and several more or less vertically oriented axial elements. One early nodule is labeled (arrow) and two other early nodules are not labeled (arrowhead). Notice that both labeled and unlabeled nodules are variable in size (compare Fig. 4 A–D). (E) Portions of four SCs from an early pachytene nucleus. One early nodule is labeled (arrow), and one late nodule is not labeled (large arrowhead). The latter nodule was identified as a late nodule because of the following combination of characteristics: association with the central region of the SC, the dense stain, and ellipsoidal shape (50 × 100 nm). (Bar = 100 nm.)
We observed 223 early nodules associated with zygotene SCs, of which almost half were labeled (97 labeled nodules/223 total nodules = 44%). By early pachynema, the majority of early nodules were labeled (26 labeled nodules/33 total nodules = 79%). Labeled and unlabeled early nodules exhibit no obvious morphological differences and appear to have approximately the same range of sizes (compare Fig. 4 A–D). In addition to early nodules, a few late nodules (n = 8) were identified on early pachytene SCs based on their higher staining density, size (50 × 100 nm), and association only with the central region of SCs. However, none of these late nodules was labeled (Fig. 4E). Because of the low sample size, these results do not exclude the presence of Rad51 in late nodules.
DISCUSSION
The pattern of numerous fluorescent Rad51 foci that we observed in leptotene, zygotene, and early pachytene nuclei from lily is similar to the results reported earlier for yeast, lily, and mouse (31–33). This pattern of fluorescent foci is also consistent with the known distribution of early nodules in plants during these early prophase I stages (6). Indeed, in spreads of lily zygotene and pachytene SCs prepared for electron microscopy, we identified many early nodules based on their frequency, variability in size, staining characteristics, and association with axial elements and SCs, and a substantial fraction of these nodules were labeled with antibodies to full-length Rad51 protein. Thus, our immunogold-labeling results demonstrate that Rad51 and/or Lim15 are components of some early meiotic nodules. Similar results regarding the labeling of Rad51 protein in early nodules have been obtained recently by Zickler (personal communication) in Sordaria and Moens et al. (41) in mouse.
Although many of the early nodules that we observed associated with zygotene and early pachytene SCs were labeled, not all early nodules were labeled. It is unlikely that this result is due to variable steric interference by chromatin because most of the chromatin was removed by DNase digestion, and SCs from the same experiment were uniformly labeled with an antibody to axial (lateral) element proteins (not shown). Early nodules can occur on either side of the central element of SCs (6), so it is possible that some of the SC-associated early nodules were not labeled because they were below an SC and inaccessible to the antibodies. However, we also observed early nodules not covered by SCs that were not labeled (see Fig. 4A). Two other possible explanations that are not mutually exclusive are: (i) There are at least two types of early nodules, and Rad51 and Lim15 are present or accessible to antibodies in only one type of nodule. (ii) Rad51 and Lim15 in early nodules are present or accessible to antibodies only transiently, thereby giving the impression of two populations of early nodules. Such a temporal alteration in the composition and/or structure of early nodules may indicate a change in the function of nodules through prophase I.
Rad51 and/or Lim15 are components of multiprotein complexes that are ultrastructurally recognizable as early meiotic nodules. Formally, it cannot be excluded that Rad51 is simply stored in early nodules. However, the dense gold-labeling of early nodules from lily is consistent with biochemical evidence that many Rad51 molecules are required to form a nucleoprotein filament that is necessary to achieve DNA pairing and strand exchange in yeast (24–27). Many proteins from the RAD52 epistasis group in yeast physically interact with one another based on in vitro (biochemical) or in vivo (yeast two-hybrid analysis) results. For example, Rad51 interacts with itself and with Rad52, Rad54, and Rad55; Rad52 interacts with itself and with Rad51; Rad55 interacts with Rad57; and Mre11 interacts with Rad50 and Xrs2 (20, 42–46). In addition, Rad51 colocalizes with Dmc1 by light microscopic analysis (31), and Rad52 interacts with Rfa1, a protein subunit of the yeast RPA single-stranded DNA-binding protein complex (47). Furthermore, immunofluorescent results show that Rad51 colocalizes with Lim15 in lily (32), and Rad51 colocalizes with RPA in mouse (48). Such observations have led many of these investigators to suggest that the proteins of the Rad52 epistasis group function together and with other proteins as multiprotein complexes in DNA repair and recombination (20, 30–33, 42–48). Given our results, any protein that colocalizes with Rad51 by immunofluorescence or that physically interacts with Rad51 protein is a possible component of early nodules also.
The ultrastructural localization of Rad51 and/or Lim15 in early meiotic nodules links the following observations: (i) numerous early meiotic nodules are associated with axial elements and SCs at early prophase I in a variety of organisms (3–6), (ii) Rad51 plays an essential role in meiotic recombination in yeast (18–20), and (iii) Rad51 is localized in numerous fluorescent foci in early prophase I nuclei from yeast, lily, and mouse (31–33). This link provides strong support for the validity of two hypotheses, namely that Rad51 has a role in meiotic recombination in higher eukaryotes and that early nodules are involved in recombination-related events.
Acknowledgments
We thank M. van Aalderen and H. Verhaar for expert technical assistance, M. A. W. Peters (Animal Facilities, Wageningen Agricultural University, The Netherlands) for immunizations, Lee Riddle at the Pacific Bulb Grower’s Research Station for providing lily bulbs, and S. Stack (Colorado State University, Fort Collins) for additional financial support. We also thank S. Stack, D. Zickler, and T. Ashley for helpful comments on the manuscript. The yeast DMC1 probe was a generous gift of D. Bishop and N. Kleckner (Harvard University, Cambridge, MA), and the antibody to lily Lim15 protein was graciously provided by T. Ogawa (National Institute of Genetics, Shizuoka, Japan). This material is based on work supported by the North Atlantic Treaty Organization under a grant awarded in 1994 to L.K.A. Additional support was provided by a Visiting Scientist grant from Wageningen Agricultural University, Grant 699 from the Colorado Agricultural Experiment Station, and Grant 95-37300-1570 from the United States Department of Agriculture to Stephen Stack.
ABBREVIATIONS
- SC
synaptonemal complex
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Moses M. Annu Rev Genet. 1968;2:363–412. [Google Scholar]
- 2.Gillies C B. CRC Crit Rev Plant Sci. 1984;2:81–116. [Google Scholar]
- 3.von Wettstein D, Rasmussen S W, Holm P B. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter A T C. Proc Natl Acad Sci USA. 1975;72:3186–3189. doi: 10.1073/pnas.72.8.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter A T C. In: Genetic Recombination. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 529–548. [Google Scholar]
- 6.Stack S M, Sherman J D, Anderson L K, Herickhoff L S. In: Chromosomes Today. Sumner A T, Chandley A C, editors. Vol. 11. London: Chapman & Hall; 1993. pp. 301–311. [Google Scholar]
- 7.Albini S M, Jones G H. Chromosoma. 1987;95:324–338. [Google Scholar]
- 8.Anderson L K, Stack S M. Chromosoma. 1988;97:96–100. doi: 10.1007/BF00286917. [DOI] [PubMed] [Google Scholar]
- 9.Zickler D, Moreau P J F, Huynh A D, Slezec A. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter A T C. Chromosoma. 1981;83:59–80. doi: 10.1007/BF00286016. [DOI] [PubMed] [Google Scholar]
- 11.Zickler D. Chromosoma. 1977;61:289–316. doi: 10.1007/BF00288615. [DOI] [PubMed] [Google Scholar]
- 12.Bernelot-Moens C, Moens P B. Chromosoma. 1986;93:220–226. [Google Scholar]
- 13.Stack S M, Anderson L K, Sherman J D. Genome. 1989;32:486–498. [Google Scholar]
- 14.Sherman J D, Stack S M. Genetics. 1995;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stack S M, Anderson L K. Chromosoma. 1986;94:253–258. [Google Scholar]
- 16.Smithies O, Powers P A. Philos Trans R Soc London B. 1986;312:291–302. doi: 10.1098/rstb.1986.0008. [DOI] [PubMed] [Google Scholar]
- 17.Carpenter A T C. Bioessays. 1987;6:232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- 18.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Pringle J R, Jones E W, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–521. [Google Scholar]
- 19.Game J C. Semin Cancer Biol. 1996;4:73–83. [PubMed] [Google Scholar]
- 20.Shinohara A, Ogawa T. Trends Biochem Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman E H, Ogawa H. Cold Spring Harbor Symp Quant Biol. 1993;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- 22.Heyer W. Experientia. 1994;50:223–233. doi: 10.1007/BF01924005. [DOI] [PubMed] [Google Scholar]
- 23.West S C. Annu Rev Biochem. 1992;61:603–640. doi: 10.1146/annurev.bi.61.070192.003131. [DOI] [PubMed] [Google Scholar]
- 24.Story R M, Bishop D K, Kleckner N, Steitz T A. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 26.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 27.Sung P, Robberson D L. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 28.Yeager Stassen N, Logsdon J M, Vora G J, Offenberg H H, Palmer J D, Zolan M E. Curr Genet. 1997;31:144–157. doi: 10.1007/s002940050189. [DOI] [PubMed] [Google Scholar]
- 29.Morita T, Yoshimura Y, Yamamoto A, Murata K, Mori M, Yamamoto H, Matsushiro A. Proc Natl Acad Sci USA. 1993;90:6577–6580. doi: 10.1073/pnas.90.14.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rockmill B, Sym M, Scherthan H, Roeder G S. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 31.Bishop D K. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 32.Terasawa M, Shinohara A, Hotta Y, Ogawa H, Ogawa T. Genes Dev. 1995;9:925–934. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]
- 33.Ashley T, Plug A W, Xu J, Solari A J, Reddy G, Golub E I, Ward D C. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- 34.Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 35.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Dunn S D. Anal Biochem. 1986;157:144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- 37.O’Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 38.Lammers J H M, van Aalderen M, Peters A H F M, van Pelt A A M, Gaemers I C, de Rooij D G, de Boer P, Offenberg H H, Heyting C. Chromosoma. 1995;104:154–163. doi: 10.1007/BF00352179. [DOI] [PubMed] [Google Scholar]
- 39.Anderson L K, Stack S M, Todd R J, Ellis R P. Chromosoma. 1994;103:357–367. doi: 10.1007/BF00417884. [DOI] [PubMed] [Google Scholar]
- 40.Griffiths G, Hoppeler H. J Histochem Cytochem. 1986;34:1389–1398. doi: 10.1177/34.11.3534077. [DOI] [PubMed] [Google Scholar]
- 41.Moens, P. B., Chen, D. J., Shen, Z., Kolas, N., Tarsounas, M., Heng, H. H. Q. & Spyropoulos, B. (1997) Chromosoma, in press. [DOI] [PubMed]
- 42.Donovan J W, Milne G T, Weaver D T. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 43.Hays S L, Firmenich A A, Berg P. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson R D, Symington L S. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johzuka K, Ogawa H. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara N, Ivanov E L, Fishman-Lobell J, Ray B L, Wu X, Haber J E. Nature (London) 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 47.Firmenich A A, Elias-Arnanz M, Berg P. Mol Cell Biol. 1995;15:1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashley, T. & Plug, A. W. in Meiosis: Current Topics in Developmental Biology, ed. Handel, M. A., in press. [DOI] [PubMed]